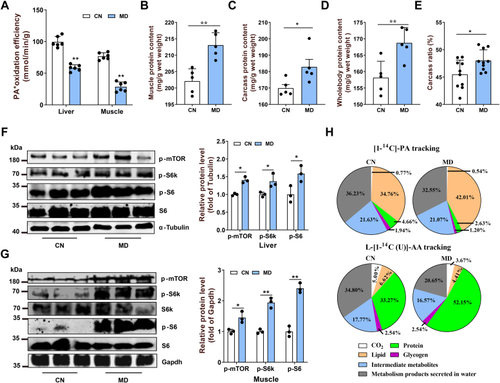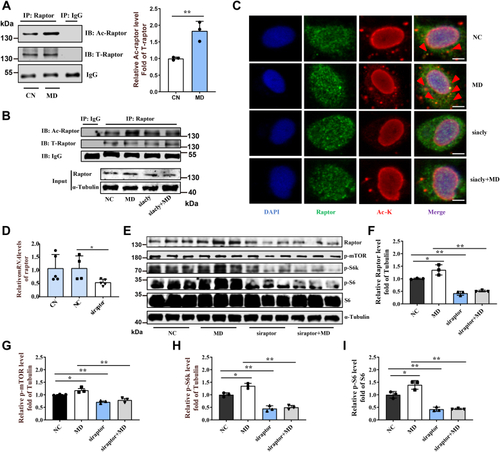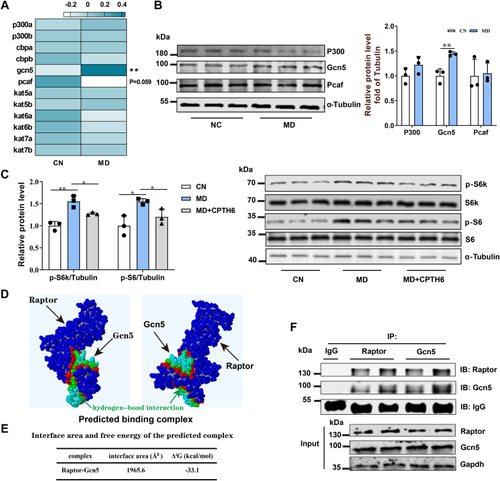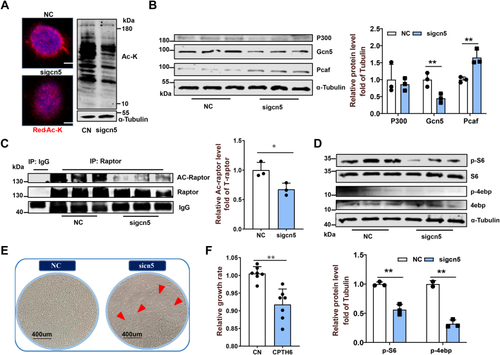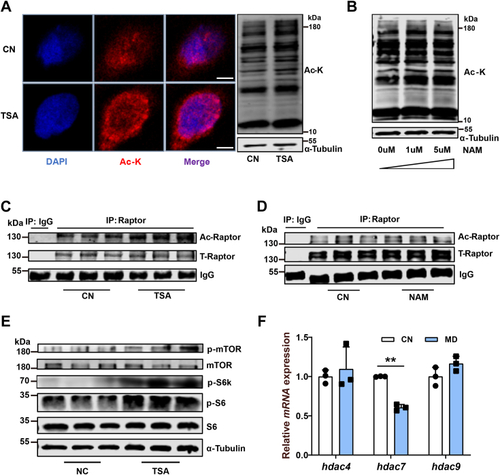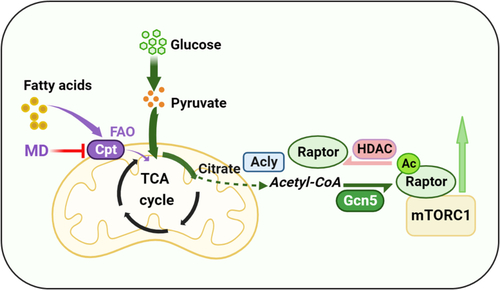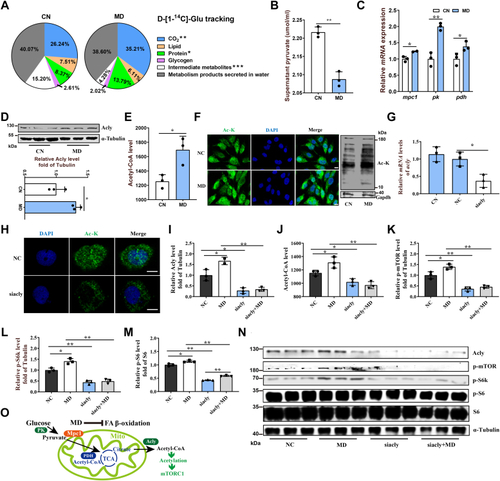
Mitochondrial FAO inhibition increases cytoplasmic glucose-derived acetyl-CoA levels and protein acetylation–mediated mTORC1 activation.A, metabolic tracking of d-[1–14C] glucose in zebrafish-fed CN and MD diets for 6 weeks (H). N = 6. B, the pyruvate content in ZFL cell culture supernatant. N = 3. The concentration and time of MD-treated cells were 1 mM and 48 h (the same below). C, relative mRNA levels of key regulatory genes (mpc1, pk [pyruvate kinase] and, pdh [pyruvate dehydrogenase]) for glucose-derived acetyl-CoA production in ZFL cells. N = 3. D and E, the protein expression levels of Acly (D; N = 3) and intracellular acetyl-CoA levels (E; N = 3) in ZFL cells. F, immunofluorescence (left) and Western blotting (right) of the global protein lysine acetylation in ZFL cells. Ac-K indicates protein lysine acetylation. Scale bars represent 10 μm. N = 3. G, relative mRNA levels of acly (ATP citrate lyase) in CN, NC, and acly siRNA-treated cells. N = 3. H, immunofluorescence of the global protein lysine acetylation in acly siRNA-treated ZFL cells. Scale bars represent 5 μm. N = 3. I, the protein expression levels of Acly were quantified after NC and MD cells were treated with acly siRNA for 48 h. N = 3. J, the intracellular acetyl-CoA levels were quantified after NC and MD cells were treated with acly siRNA for 48 h. N = 3. K–M, the effect of acly knockdown on mTORC1 signaling caused by mitochondrial FAO inhibition. NC and MD cells were treated with acly siRNA for 48 h. Relative protein quantification of p-mTOR/tubulin (K), p-S6k/tubulin (L), and p-S6/S6 (M) in ZFL cells. N = 3. N, the Western blots for Acly, p-mTORSer2448, p-S6kThr389, p-S6Ser235/236, and S6 are shown. N = 3. O, mitochondrial FAO inhibition increases glucose-derived acetyl-CoA production, which in turn promotes cytoplasmic protein acetylation-mediated mTORC1 activation. Data represent mean ± SD. ∗p < 0.05 and ∗∗p < 0.01. Acly, ATP citrate lyase; CN, control; FAO, fatty acid oxidation; MD, mildronate; mTORC1, mechanistic target of rapamycin complex 1; NC, negative control; ZFL, zebrafish liver.
|