- Title
-
Cell competition for neuron-derived trophic factor controls the turnover and lifespan of microglia
- Authors
- Yu, T., Kuang, H., Wu, X., Huang, Y., Wang, J., Wen, Z.
- Source
- Full text @ Sci Adv
|
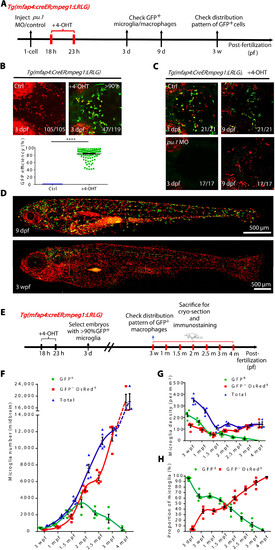
The developmental dynamics of RBI and AGM microglia. (A) A timeline indicates 4-OHT treatment and fluorescent imaging of Tg(mfap4:CreER;mpeg1:LRLG) embryos. (B) Fluorescence imaging of microglia in the brain of 3 dpf Tg(mfap4:CreER;mpeg1:LRLG) embryos treated with 4-OHT from 18 to 23 hpf (top) and quantification of the label efficiency by percentage (GFP+ cells in total microglia in 3-dpf brain) (bottom) (Ctrl n = 105, +4-OHT n = 119). (C) Fluorescence imaging of microglia in the brain of 3 and 9 dpf control or pu.1 MO injected Tg(mfap4:CreER;mpeg1:LRLG) embryos treated with 4-OHT from 18 to 23 hpf. (D) Fluorescence imaging of 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) larva at 9 dpf and 3 wpf. Zebrafish larvae were in lateral view with anterior to the left to visualize the distribution of GFP+ cells throughout the body. (E) Schematic diagram shows the experimental setup for the treatment of embryos with 4-OHT and collection of samples for cryo-section. (F) Quantification of the absolute number of RBI and AGM microglia in the developing and adult midbrain. (G) Quantification of the density of RBI and AGM microglia in the developing and adult midbrain. (H) Quantification of the proportion of RBI and AGM microglia in the developing and adult midbrain. (F) to (H) Data were presented as mean ± SEM (3 dpf n = 12, 3 wpf n = 5, 1 mpf n = 5, 1.5 mpf n = 5, 2 mpf n = 5, 2.5 mpf n = 5, 3 mpf n = 4, and 4 mpf n = 3). |
|
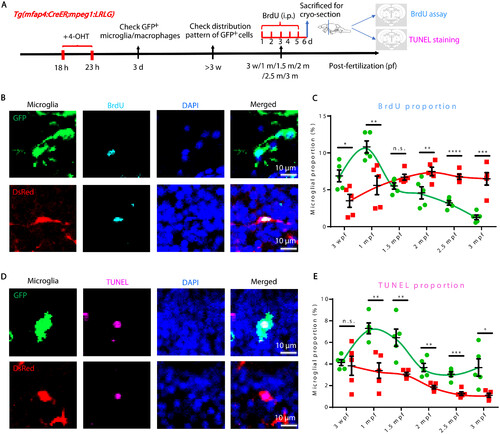
BrdU and TUNEL assays reveal different proliferation and death rates of RBI and AGM microglia. (A) Schematic diagram shows the experimental setup for BrdU and TUNEL assays. (B) Representative images of GFP+ RBI microglia and GFP−DsRed+ AGM microglia costained with BrdU and DAPI. (C) Quantification of the proliferation rate of RBI (BrdU+GFP+/GFP+) and AGM (BrdU+GFP−DsRed+/GFP−DsRed+) microglia. (D) Representative images of GFP+ RBI microglia and GFP−DsRed+ AGM microglia costained with TUNEL and DAPI. (E) Quantification of the death rate of RBI (TUNEL+GFP+/GFP+) and AGM (TUNEL+GFP−DsRed+/GFP−DsRed+) microglia. not significant (n.s.), P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (Student’s t test). Data were presented as mean ± SEM (n = 5). i.p., intraperitoneal. |
|
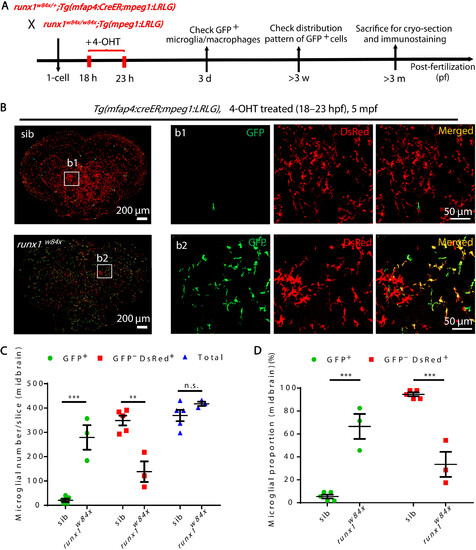
Expansion and prolonged lifespan of RBI microglia in AGM microglia-deficient runx1w84x mutants. (A) Schematic diagram shows the experimental setup for the tracing of RBI microglia in siblings and runx1w84x mutants. (B) Representative images of the midbrain cross section of 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) siblings and runx1w84x;Tg(mfap4:CreER;mpeg1:LRLG) mutant fish at 5 mpf. GFP+ and GFP−DsRed+ cells represent RBI and AGM microglia, respectively. (C and D) Quantification of the number (C) and proportion (D) of RBI (GFP+) and AGM (GFP−DsRed+) microglia in (B). n.s., P > 0.05; **P < 0.01; ***P < 0.001 (Student’s t test). Data were presented as mean ± SEM (sibling n = 5, runx1w84x n = 3). |
|
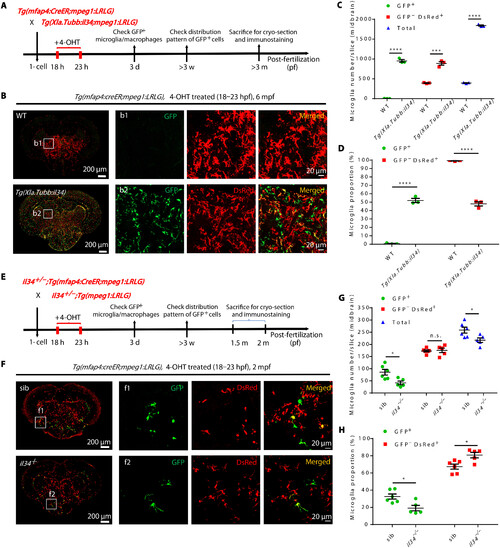
Alteration of neuron-derived Il34 level changes the proportion and lifespan of RBI microglia. (A) Schematic diagram shows the experimental setup for RBI microglia tracing in WT and Tg(Xla.Tubb:il34) fish. (B) Representative images of the midbrain cross section of 4-OHT–treated Tg(mfap4:creER;mpeg1:LRLG) and Tg(Xla.Tubb:il34;mfap4:creER;mpeg1:LRLG) fish at 6 mpf. GFP+ and GFP−DsRed+ cells represent RBI and AGM microglia, respectively. (C and D) Quantification of the number (C) and proportion (D) of RBI (GFP+) and AGM (GFP−DsRed+) microglia in (B) (n = 3 for each genotype). (E) Schematic diagram shows the experimental setup for RBI microglia tracing in siblings and il34−/− mutants. (F) Representative images of the midbrain cross section of 4-OHT–treated Tg(mfap4:CreER;mpeg1:LRLG) siblings and il34−/−;Tg(mfap4:CreER;mpeg1:LRLG) mutant fish at 2 mpf. GFP+ and GFP−DsRed+ cells represent RBI and AGM microglia, respectively. (G and H) Quantification of the number (G) and proportion (H) of RBI (GFP+) and AGM (GFP−DsRed+) microglia in (F) (sibling n = 6, il34−/− n = 5). n.s., P > 0.05; *P < 0.05; ***P < 0.001; ****P < 0.0001 (Student’s t test). Data were presented as mean ± SEM. |
|
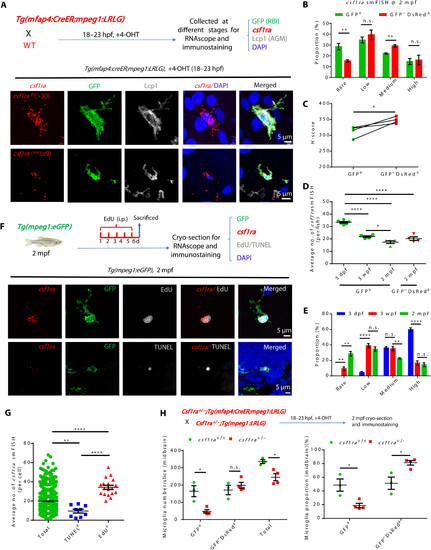
Age-dependent decline of csf1ra expression accounts for the less competitiveness of RBI microglia. (A) Diagram shows the design for RNAscope smFISH (red), anti-GFP (green), anti-Lcp1 (white), and DAPI (blue) staining (top) and the representative images of RBI microglia with high and rare csf1ra expression (bottom). (B) Quantification of four categories of RBI and AGM microglia expressing different csf1ra levels at 2 mpf (n = 4). (C) H-score quantification of csf1ra expression levels in RBI and AGM microglia (n = 4). (D) Average number of csf1ra smFISH dots in 3-dpf, 3-wpf, and 2-mpf RBI microglia and in 2-mpf AGM microglia (3 dpf n = 6, 3 wpf n = 4, 2 mpf n = 4). (E) Quantification of four categories of RBI microglia expressing different csf1ra level (3 dpf n = 6, 3 wpf n = 4, 2 mpf n = 4). (F) Diagram shows the experimental setup for RNAscope smFISH (red), anti-GFP (green), EdU or TUNEL (white), and DAPI (blue) staining (top) at 2 mpf, and representative images show the expression of csf1ra in EdU+GFP+ and TUNEL+GFP+ microglia at 2 mpf (bottom). (G) Quantification of csf1ra smFISH dots in EdU+GFP+, TUNEL+GFP+, and GFP+ (total) microglia in (F) (total n = 1255, TUNEL+ n = 9, EdU+ n = 18). (H) Diagram shows the design for RBI microglia tracing in WT, csf1ra+/−, and csf1ra−/− mutants (top) and quantification of the number and proportion of RBI (GFP+) and AGM (GFP−DsRed+) microglia in WT and csf1ra+/− fish at 2 mpf (WT n = 3, csf1ra+/− n = 4). n.s., P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001 (Student’s t test). Data were presented as mean ± SEM. |
|
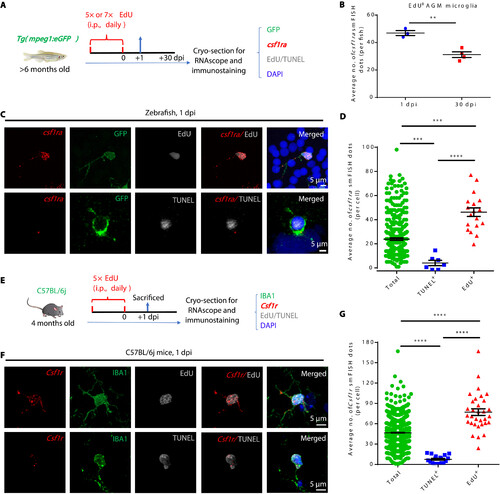
Csf1ra/CSF1R level determines the fitness of adult microglia in zebrafish and mice. (A) Schematic diagram shows the experimental setup for RNAscope smFISH (red), anti-GFP/Lcp1 (green), EdU or TUNEL (white), and DAPI (blue) multicolor staining in adult zebrafish. (B) Quantification of csf1ra smFISH dots in EdU+GFP+ at 1 dpi and 30 dpi (1 dpi n = 3, 30 dpi n = 4). (C) Representative images show the expression of csf1ra in EdU+GFP+ and TUNEL+GFP+ microglia in adult zebrafish. (D) Quantification of csf1ra smFISH dots in EdU+GFP+, TUNEL+GFP+, and GFP+ (total) microglia at 1 dpi (total n = 312, TUNEL+ n = 7, EdU+ n = 18). (E) Schematic diagram shows the experimental setup for RNAscope smFISH (red), anti-IBA1 (green), EdU or TUNEL (white), and DAPI (blue) multicolor staining in adult mice. (F) Representative images show the expression of Csf1r in EdU+IBA1+ and TUNEL+IBA1+ microglia in 4-month-old C57BL/6j mice. (G) Quantification of csf1r smFISH dots in EdU+IBA1+, TUNEL+IBA1+, and IBA1+ (total) microglia shown in (F) (total n = 1348, TUNEL+ n = 18, EdU+ n = 32). **P < 0.01; ***P < 0.001; ****P < 0.0001 (Student’s t test). Data were presented as mean ± SEM. |