- Title
-
Social and asocial learning in zebrafish are encoded by a shared brain network that is differentially modulated by local activation
- Authors
- Pinho, J.S., Cunliffe, V., Kareklas, K., Petri, G., Oliveira, R.F.
- Source
- Full text @ Commun Biol
|
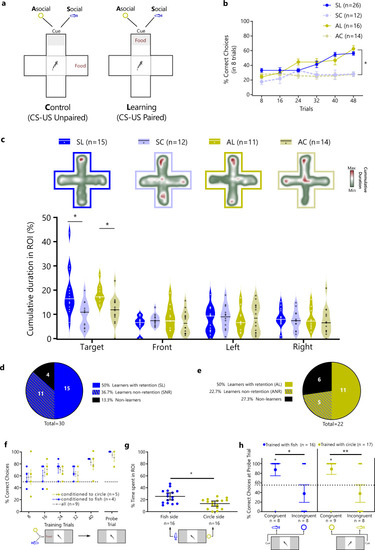
Social and asocial classic conditioning in zebrafish. |
|
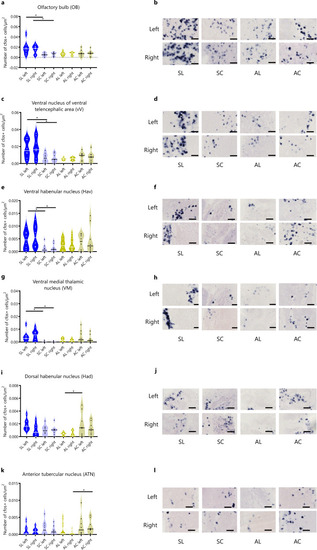
Neuronal activity associated with social (a–h) and asocial (i–l) classic conditioning in zebrafish assessed by in situ hybridization of the immediate early gene Representative photomicrographs of |
|
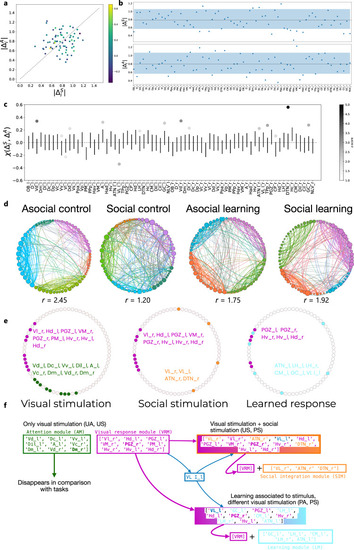
Brain networks associated with social and asocial learning. |