- Title
-
Zebrafish ambra1b knockout reveals a novel role for Ambra1 in primordial germ cells survival, sex differentiation and reproduction
- Authors
- Fontana, C.M., Terrin, F., Facchinello, N., Meneghetti, G., Dinarello, A., Gambarotto, L., Zuccarotto, A., Caichiolo, M., Brocca, G., Verin, R., Nazio, F., Carnevali, O., Cecconi, F., Bonaldo, P., Dalla Valle, L.
- Source
- Full text @ Biol. Res.
|
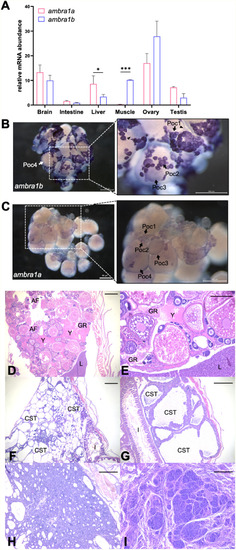
Expression patterns of ambra1 mRNAs and pathological findings in the zebrafish ambra1a and ambra1b KO lines. (A) RT-qPCR analysis of ambra1a and ambra1b mRNA expression in different adult zebrafish tissues. Data were generated from different biological replicates, each consisting of tissue samples from single individual (intestine, liver, ovary n = 4; brain, muscle n = 3; testis n = 2; intestine, liver, brain and muscle were a mix of female and male tissues). qPCR data were analyzed by 2−∆CT method and actb2 was selected as reference gene for normalization. Values represent the mean ± SEM. Statistical analysis was performed using Student’s t-test. * P < 0.05; *** P < 0.001. (B, C) Whole mount in situ hybridization with ambra1b (B) and ambra1a probes (C) in ovaries of 6-month-old WT zebrafish. Arrowheads point at somatic cells surrounding oocytes. Poc1, primary oocytes stage 1; Poc2, primary oocytes stage 2; Poc3, previtellogenic oocytes; Poc4, vitellogenic oocytes. (D) Representative ovary of a 18-mpf ambra1a−/− female showing follicular degeneration and presence of yolk free in the coelomic cavity as a result of degenerated follicles (H&E; scale bar, 200 μm). (E) Higher magnification of panel D, showing a moderate granulomatous reaction towards proteinaceous material (H&E; scale bar, 400 μm). (F) Testis of a 20-mpf ambra1b−/− male, showing diffuse cystic degeneration of the coelomic cavity (H&E; scale bar, 400 μm). (G) Testis of a 15-mpf ambra1b−/− male, showing diffuse ectasia of seminiferous tubules and hyperplasia of spermatogonia lining the tubules (H&E; scale bar, 200 μm). (H) Testis of 18-mpf ambra1a−/− male, displaying seminoma (H&E; scale bar, 200 μm). (I) Testis of a 17-mpf ambra1b−/− male, containing an undifferentiated germ cell tumor (H&E; scale bar, 200 μm). AF, atretic follicle; CST, cystic seminiferous tubule; GR, granulomatous reaction; I, intestine; L, liver; Y, yolk. |
|
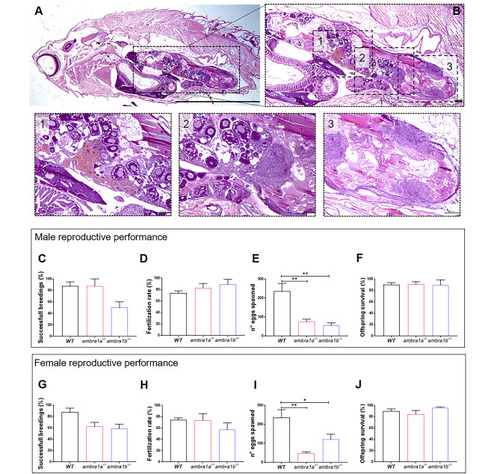
KO of ambra1a and ambra1b reduces the reproductive capabilities of zebrafish in both sexes. (A) Representative histological analysis of 16-mpf ambra1b−/− female, showing H&E staining of total body sagittal section. Scale bar, 5 mm. (B) Magnification of the dotted area of panel A, showing the ovary region. Scale bar, 400 μm. The lower panels show higher magnifications of the respective dotted areas (1–3), corresponding to different regions of the ovary. Scale bar, 200 μm. (C-J) Quantification of the reproductive performance of WT, ambra1a−/− and ambra1b−/− males (C-F) and females (G-J). Panels C and G show the percentage of spawning success in mutant male and female tests, respectively. Panels D and H show the percentage of fertilized eggs on the total number of eggs. Panels E and I show the mean number of eggs spawned in successful breeding events. Panels F and J show the percentage of survival of the offspring at 5 dpf. Males: ambra1a−/− n = 4; ambra1b−/− n = 4; WT n = 4. Females: ambra1a−/− n = 4; ambra1b−/− n = 3; WT n = 4. Error bars indicate SEM. Statistical analysis was performed using One way ANOVA. * P < 0.05; ** P < 0.01 PHENOTYPE:
|
|
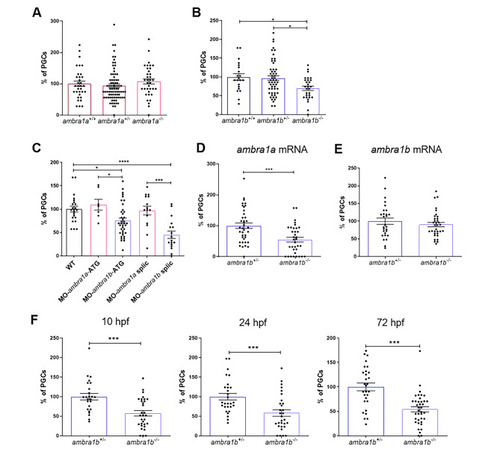
KO of ambra1b leads to decreased PGC number. (A, B) Quantification of PGCs in 10-hpf embryos of ambra1a (A), ambra1b (B) genotypes, expressed as percentage of PGCs in heterozygous and homozygous siblings compared to their WT controls. Histograms show data pooled together from three independent experiments. Only ambra1b−/− embryos show significant reduction of PGC number (ambra1a+/+ n = 36, ambra1a+/− n = 78, ambra1a−/− n = 35; ambra1b+/+ n = 21, ambra1b+/− n = 54, ambra1b−/− n = 29). Statistical analysis was performed using One way ANOVA. *, P < 0.05. (C) Percentage of PGCs in WT embryos injected with ambra1a or ambra1b ATG MOs or splicing MOs. Analysis was performed at 10 hpf by immunohistochemistry for Vasa protein. ambra1b knockdown performed with both types of MO results in a significant reduction of PGCs. Histograms show data pooled together from three independent experiments (WT n = 23, MO-ambra1a-ATG n = 14, MO-ambra1b-ATG n = 41, MO-ambra1a-splic n = 20, MO-ambra1b-splic n = 20). Statistical analysis was performed using One way ANOVA. *, P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. (D, E) Results of rescue experiments by co-injection of GFP-nos1-3’UTR mRNA with ambra1a or ambra1b mRNA, respectively. PGC number is expressed as percentage with respect to ambra1b+/− animals, in which the PGC number was settled as 100%. Histograms show data pooled together from three independent experiments (D: ambra1b+/− n = 38, ambra1b−/− n = 35; E: ambra1b+/− n = 28, ambra1b−/− n = 34). Statistical analysis was performed using Student’s t-test. *** P < 0.001. (F) Quantification of PGC number in ambra1b+/− and ambra1b−/− embryos at 10, 24,72 hpf after injection with GFP-nos1-3’UTR mRNA. Number of PGCs in controls (ambra1b+/−) was settled as 100%. Histograms show data pooled together from three independent experiments (10 hpf: ambra1b+/− n = 25, ambra1b−/− n = 31; 24 hpf: ambra1b+/− n = 28, ambra1b−/− n = 30; 72 hpf: ambra1b+/− n = 29, ambra1b−/− n = 38). Statistical analysis was performed using Student’s t-test. *** P < 0.001. Error bars indicate SEM. PHENOTYPE:
|
|
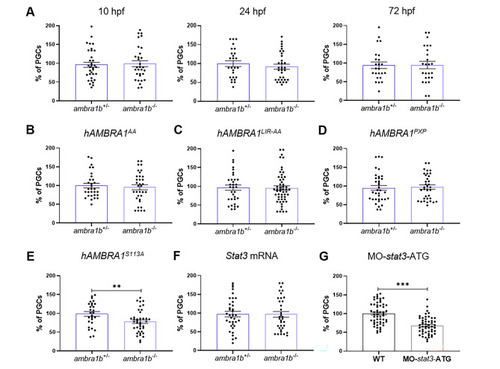
Injection of hAMBRA1 mRNA counteracts the loss of PGCs. (A) Quantification of PGCs at 10, 24 and 72 hpf after co-injection of one-cell embryos with GFP-nos1-3’UTR and hAMBRA1 mRNA (10 hpf: ambra1b+/− n = 34, ambra1b−/− n = 31; 24 hpf: ambra1b+/− n = 26, ambra1b−/− n = 32; 72 hpf: ambra1b+/− n = 24, ambra1b−/− n = 25). (B-E) Quantification of PGCs after injection with GFP-nos1-3’UTR mRNA and hAMBRA1 mRNA mutated in TRAF6 (hAMBRA1AA, B), LC3 (hAMBRA1LIR − AA, C), PP2A (hAMBRA1PXP, D) and CUL4–DDB1 (hAMBRA1S113A, E) binding sites (B: ambra1b+/− n = 30, ambra1b−/− n = 34; C: ambra1b+/− n = 33, ambra1b−/− n = 54; D: ambra1b+/− n = 36, ambra1b−/− n = 35; E: ambra1b+/− n = 28, ambra1b−/− n = 39). (F) Injection of one-cell stage ambra1b+/− and ambra1b−/− embryos with murine Stat3 mRNA and quantification of PGCs at 10-hpf (ambra1b+/− n = 29, ambra1b−/− n = 27). (G) Injection of one-cell stage WT embryos with MO-stat3-ATG and quantification of PGCs at 10-hpf (ambra1b+/− n = 55, ambra1b−/− n = 52). In all panels, the PGC number is expressed as percentage with respect to ambra1b+/− animals, in which the PGC number was settled as 100%. The histograms show data pooled together from four (A, F) or three (B, C, D, E, G) independent experiments. Error bars indicate SEM. Statistical analysis was performed using Student’s t-test. ** P < 0.01, *** P < 0.001 PHENOTYPE:
|
|
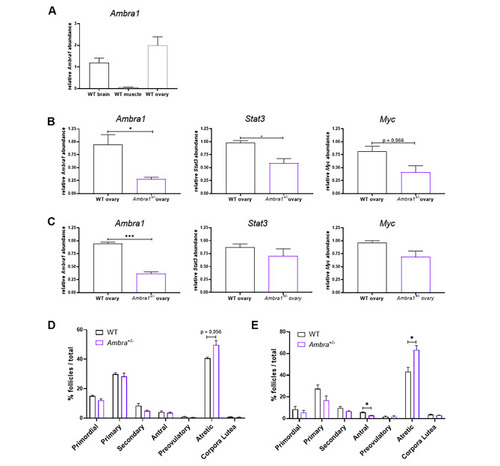
Ambra1 deficiency affects ovary physiology in mice. (A) RT-qPCR analysis of Ambra1 mRNA levels in brain, muscle and ovaries of 3-month-old WT mice. (B, C) RT-qPCR analysis of Ambra1, Stat3 and Myc expression in 3-month-old (B) and 10-month-old (C) Ambra1+/+ (WT) and Ambra1+/− sibling mice. Data were generated from three mice of each genotype for each time point. Values represent the mean ± SEM. Data were normalized with the housekeeping gene Gapdh. Statistical analysis was performed using Student’s t-test. *, P < 0.05. (D, E) Comparison of follicle proportion in ovaries of 3-month-old (D) and 10-month-old (E) Ambra1+/+ and Ambra1+/− mice. The proportion of follicles is displayed as the percentage of each follicle type over the total number of follicles (primordial, primary, secondary, antral, preovulatory, atretic and corpora lutea) over the total number of follicles. Values represent the mean ± SEM. Data were generated from three mice of each genotype for each time point, except for 3-month-old Ambra1+/− mice (four ovaries). Statistical analysis was performed using Student’s t-test. *, P < 0.05 |