- Title
-
STAT5b is a key effector of NRG-1/ERBB4-mediated myocardial growth
- Authors
- Vaparanta, K., Jokilammi, A., Paatero, I., Merilahti, J.A., Heliste, J., Hemanthakumar, K.A., Kivelä, R., Alitalo, K., Taimen, P., Elenius, K.
- Source
- Full text @ EMBO Rep.
|
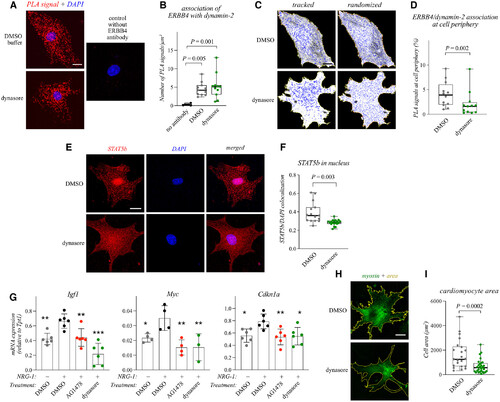
Figure 1. NRG-1 promotes cardiomyocyte hypertrophy via ERBB4 and STAT5b Data information: For all boxplots the central band represents the median, the box the interquartile range and whiskers the whole range of values. Source data are available online for this figure. |
|
Figure 2. Dynamin-2 controls ERBB4 signaling and cardiomyocyte hypertrophy Data information: For all boxplots the central band represents the median, the box the interquartile range and whiskers the whole range of values. Source data are available online for this figure. |
|
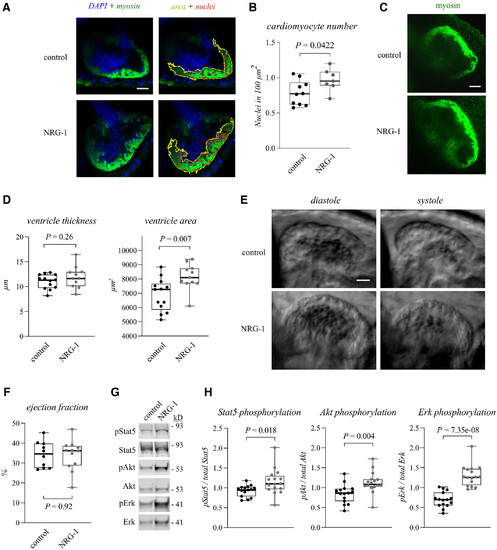
Figure 3. NRG-1 promotes hyperplastic myocardial growth and Stat5 activation in zebrafish embryos Data information: For all boxplots the central band represents the median, the box the interquartile range and whiskers the whole range of values. Source data are available online for this figure. |
|
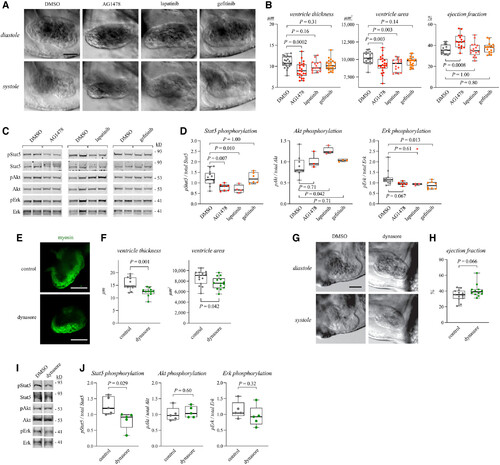
Figure 4. Erbb4 pathway regulates myocardial growth and Stat5 activation in zebrafish embryos Data information: For all boxplots the central band represents the median, the box the interquartile range and whiskers the whole range of values. Source data are available online for this figure. |
|
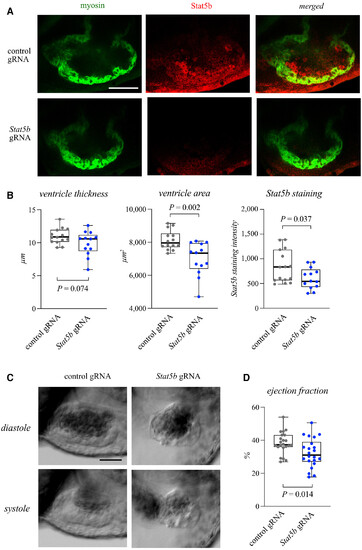
Figure 5. Stat5b is necessary for cardiac growth and function in zebrafish embryos Data information: For all boxplots the central band represents the median, the box the interquartile range and whiskers the whole range of values. Source data are available online for this figure. |
|
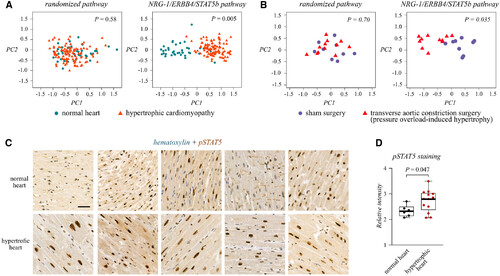
Figure 6. NRG-1/ERBB4/STAT5b signaling pathway is perturbed in pathological cardiac hypertrophy Source data are available online for this figure. |
|
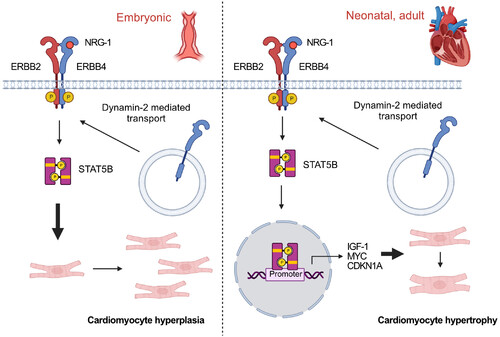
Figure 7. The proposed NRG-1/ERBB4/STAT5b pathway in cardiac growth The pathway may regulate both cardiomyocyte hyperplasia (left) and hypertrophy (right) at different developmental stages. |