Fig. 1
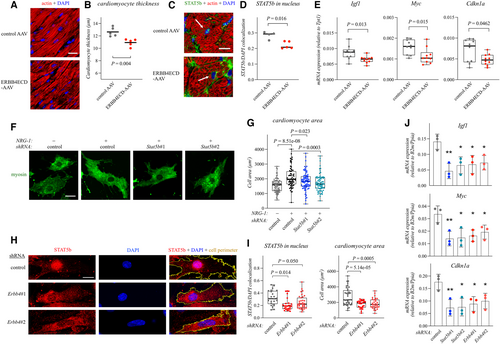
Figure 1. NRG-1 promotes cardiomyocyte hypertrophy via ERBB4 and STAT5b Data information: For all boxplots the central band represents the median, the box the interquartile range and whiskers the whole range of values. Source data are available online for this figure.
Image
Figure Caption
Source Data for Figure 1 [embr202256689-sup-0015-SDataFig1.zip]
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ EMBO Rep.