- Title
-
A small change with a twist ending - a single residue in EGF-CFC drives Bilaterian asymmetry
- Authors
- Truchado-García, M., Perry, K.J., Cavodeassi, F., Kenny, N.J., Henry, J.Q., Grande, C.
- Source
- Full text @ Mol Bio Evol
|
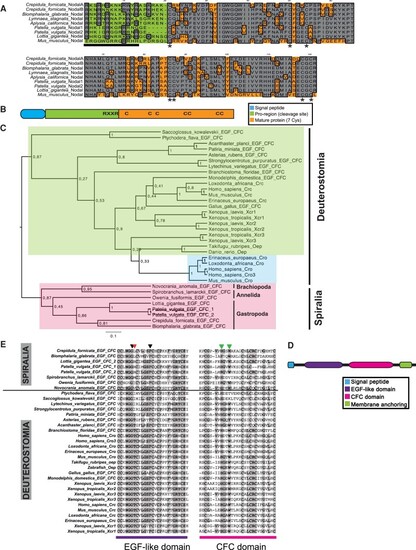
Two paralogs for Nodal are present in Crepidula fornicata, and EFG-CFC orthologs are present in non-deuterostomes. (A) Alignment of several ortholog proteins for Nodal in different gastropod species (C. fornicata; Biomphalaria glabrata; Lymnaea stagnalis; Aplysia californica; Patella vulgata; Lottia gigantea) and mammalian Mus musculus. Gray indicates conservation for one given position; asterisks point the seven conserved cysteines that characterize the family; bar delimits the regions were cleavage sites are located. (B) Generic structure of Nodal protein. C, cysteines; (RXXR), cleavage site. (C) EFG-CFC proteins inter-relationships. Bayesian phylogenetic tree of EFG-CFC inferred proteins from spiralian species, along with previously described sequences of other bilaterians (supplementary S1, Supplementary Material online). The WAG model (Whelan and Goldman 2001) was selected as the best-fit model of protein evolution using ProtTest (Abascal et al. 2005). Tree shown is the result of Bayesian analysis, run for 25,000,000 generations, and analyses run until the SD of split frequencies was below 0.01, with the first 25% of sampled trees discarded as “burn-in.” Posterior probabilities can be seen at each branch. Scale bar represents substitutions per site at given distances. Shading shows major protein lineages. (D) Generic structure of EGF-CFC protein. (E) Alignment of the EGF-like and CFC domains of several ortholog proteins for Cripto, Cryptic, and EGF-CFC in different bilaterians, from both Spiralia and Deuterostomia clades. Shading indicates conservation for one given position. Arrowheads indicate the relevant positions in the ligand-receptor-coreceptor binding; Gly and Phe (black arrows) are conserved, whereas Thr, described as necessary for Nodal signaling, is only present in chordates and some lineages of echinoderms, and is variable in spiralians (red arrow); His and Trp (green arrows) are conserved in all orthologs with the exception for Spirobranchus and hemichordates. |
|
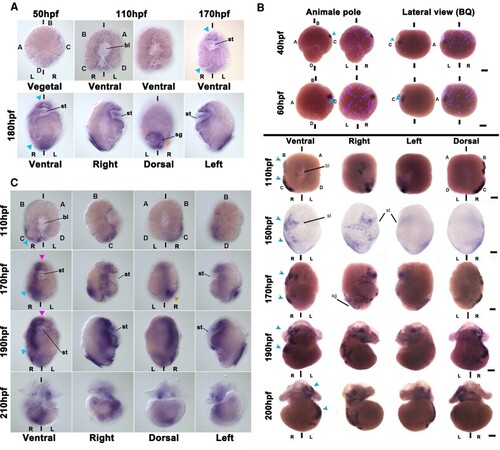
Cfo-nodalB expression suggests involvement in LRA development. (A–C) Spatio-temporal localization of (A) Cfo-nodalA, (B) Cfo-nodalB, and (C) Pitx mRNA by WISH during the development of Crepidula fornicata. Several views of the same embryo for each stage are indicated on the figure. bl, blastopore; L, left; R, right; sg, shell gland; st, stomodeum. A, B, C, and D indicate the quadrants; hpf, hours post fertilization; bilateral axis is represented by the vertical bars. Scale corresponds to 30 μm. (A) Note that the expression is symmetrical in all the stages (blue arrowheads); orange arrowhead, shell gland. In (B), superpositions of brightfield images with Hoechst staining have been included for early stages, to identify the cells. Note that the expression is asymmetrical in all the stages (blue arrowheads). (C) Pink arrowhead, symmetrical signals; blue arrowhead, asymmetrical signals; orange arrowhead, shell gland. |
|
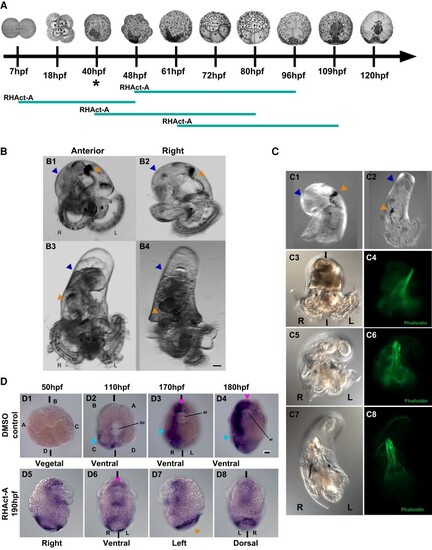
Cfo-nodalB regulates the establishment of LRA, whereas Activin inhibits the Nodal pathway in Crepidula fornicata. (A) Diagram of the windows used on RHAct-A treatment. The teal bar represents the time in which the embryos were cultured in RHAct-A, and the asterisk indicates when nodalB expression first starts; hpf, hours post fertilization. (B) Overexpression of Cfo-nodalB, in a 2-week-old hatched veliger. (B1) Anterior and (B2) right views of a control veliger; (B3) anterior and (B4) right views of a veliger after Cfo-nodalB injection. (C) RHAct-A treatment, in a 2-week-old hatched veliger. Right view of a (C1) control veliger and (C2) a treated veliger. (C3–C8) Three individuals with different grades of coiling after treatment. Brightfield of (C3) coiled DMSO control, (C5) hemitorsioned, and (C7) straight larvae after RHAct-A. (C4/C6/C8): note the change of the retractor muscle of those same larvae (C3, C5, C7) shown by phalloidin stain. In both B and C, note the bilateral symmetry phenotype in B3, B4 and C2, C7. L, left; R, right; shell coiling, blue arrowhead; gut coiling, orange arrowhead. (D) Spatio-temporal localization of Pitx mRNA by WISH, in RHAct-A treated C. fornicata embryos. (D1–D4) Control embryos in vegetal/ventral view in four stages during development. D1, non-stained blastula (50 hpf); D2, mid-gastrula (110 hpf); D3, organogenesis (170 hpf); D4, preveliger larva (180 hpf). Note the right-side staining (cyan arrowhead) and the symmetrical staining (pink arrowhead) on the anterior region and stomodeum (D2–D4). (D5–D8) Views of a preveliger, post-RHAct-A exposure. Note that the symmetrical expression remains (pink arrowhead), whereas the asymmetrical expression is lost on the right side; shell gland, orange arrowhead. st, stomodeum; bl, blastopore; L, left; R, right. A, B, C and D indicate the quadrants; bilateral axis is represented by the vertical bars. Scale bars correspond to 30 μm. |
|
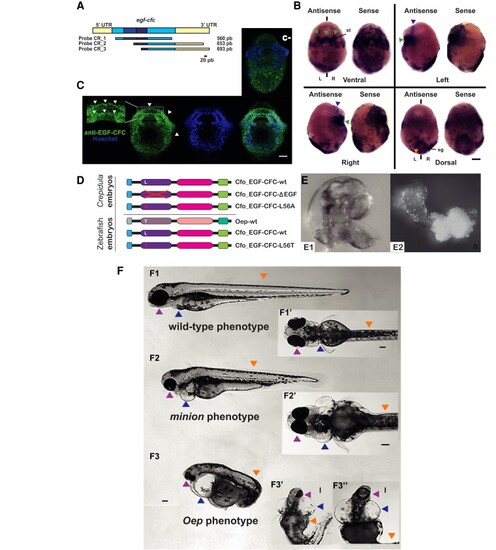
Cfo-EGF_CFC is not involved in the establishment of LRA in Crepidula fornicata, but a single substitution is able to the rescue of Zebrafish Oep mutants. (A) Cfo-egf-cfc probe cloning designs. Regions in yellow represent the 5′- and 3′-UTRs, and blue corresponds to the coding region. Note that the two darkest fragments correspond to the EGF-like and CFC domains. Probe 1 contains most part of the coding region, whereas probes 2 and 3 contain different lengths of coding region and 3′-UTR. The bar indicates the scale. (B) Localization of Cfo-egf_cfc transcripts by WISH in a C. fornicata preveliger larva with probe 1. The left embryo on each view was incubated with the antisense probe, and the right one corresponds to the sense probe. Green arrowheads point to the symmetrical signal at the stomodeum; purple arrowhead, symmetrical anterior signals; orange, apparent symmetrical shell gland signals. st, stomodeum; sg, shell gland; L, left; R, right; bilateral axis is represented by the vertical bars. Scale corresponds to 30 μm. (C) Ventral view of an immunofluorescent staining for EGF-CFC on a C. fornicata preveliger larva. Green channel shows the staining from a specific antibody designed against this protein, which signal appears to be symmetrical and consistent with the result of WISH (fig. 4BB). Magnification of the stomodeum reveals this protein in the cell membranes (white arrows). The blue channel shows nuclei after Hoechst staining; followed by the merge of both channels. An equivalent larva is provided from the negative control reaction (c−); note the lack of staining at the stomodeum. Scale corresponds to 30 μm. (D) mRNA constructs injected in Crepidula and Zebrafish embryos. (E1) Left side of a wild-type C. fornicata juvenile; (E2) radialized individual resulting from EGF-CFC LOF, after using the construct lacking the EGF-like domain. (F) Zebrafish individuals resulting from the rescue experiment on oep mutants, after injecting zygotes with substitution L56T on the EGF-CFC construct. (F1) Left and (F1′) ventral views of a wild-type phenotype; (F2) left and (F2′) ventral views of a minion phenotype; (F3) left and (F3′ and F3″) ventral views of a classic oep phenotype. Arrowheads point to the structures affected the phenotype: Pink, lens and head; blue, heart; orange, anterior-posterior axis. Scale bars correspond to 100 μm. |
|
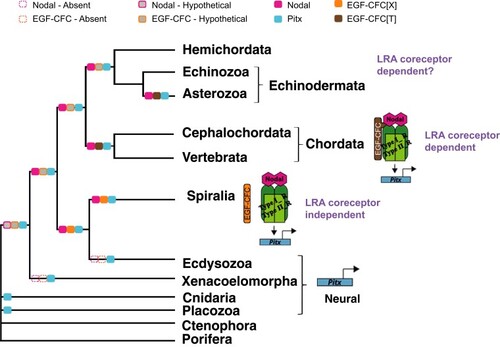
Proposed model for the evolution of EGF-CFC role in Metazoa. The orthologs for EGF-CFC in spiralian representatives display different amino acids on this relevant position in the EGF-like domain, whereas all representatives of Chordata and some echinoderms display a threonine. The change of amino acid in this specific position would have happened either in the ancestor of Chordata or in the ancestor of all deuterostomes (see supplementary S3, Supplementary Material online). This gave the coreceptor the ability to interact with the ligand, and regulate Pitx expression, thereby participating in the establishment of LRA. In Spiralia and its ancestor, the function of EGF-CFC would not be linked to LRA, but to gastrulation and axis determination. The regulation of LRA is proposed to be independent of the coreceptor for Spiralia. Pitx ancestral function would be related to neural specification, and it would remain as the only function of this gene in those clades that lack the ligand and coreceptor. The color filled boxes represent presence of the protein in a given clade. The gray filled boxes represent inferred hypothetical presence of the protein. The dotted line boxes represent the absence of an ortholog. X, any amino acid; T, threonine; LRA, left-right asymmetry. |