- Title
-
leptin b and its regeneration enhancer illustrate the regenerative features of zebrafish hearts
- Authors
- Shin, K., Begeman, I.J., Cao, J., Kang, J.
- Source
- Full text @ Dev. Dyn.
|
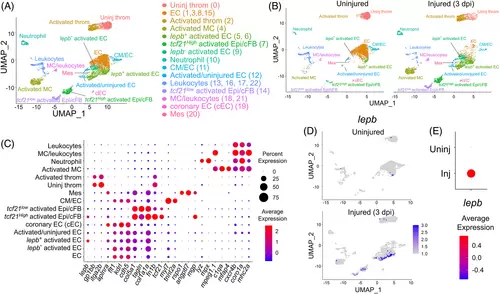
Heterogenous cell clusters in injured hearts of adult zebrafish. (A) Clustering assignments of runx1P2:Citrine or kdrl:mCherry expressing cells collected from uninjured and injured hearts. Uniform Manifold Approximation and Projection (UMAP) axes were calculated by unsupervised clustering method. throm, thrombocyte. EC, endocardial/endothelial cells. Epi, epicardial cells. cFB, cardiac fibroblasts. Mes, mesenchyme-like cells. MC, macrophages. Original cluster numbers are indicated in the Parenthesis. (B) Clustering assignments for uninjured and injured hearts. dpi, days postinjury. (C) Differential expression of the key marker genes to identify cell types shown as a dot plot. (D) Injury-dependent expression of lepb depicted by UMAP plot. (E) Injury-dependent expression of lepb depicted by a dot plot |
|
lepb marks a subset of EC population producing regenerative factors. (A) Robust induction of lepb in a subset of ECs activated by injury. (B) Differentially expressed genes between lepb+ and lepb− |
|
Heterogenous Epi/cFB in zebrafish injured hearts. (A) Injury-inducible expression of cardiac fibroblast marker genes in tcf21low and tcf21high activated Epi/cFB. (B) Enrichment of cells expressing key epicardial transcription factors (TFs) and lepb in tcf21high |
|
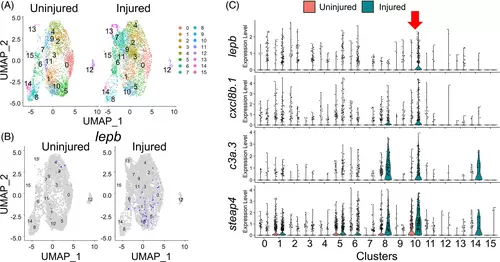
lepb marks a specific subpopulation of tcf21+ Epi/EPDCs. (A) Clustering assignments of tcf21+ Epi/EPDCs collected from uninjured and injured hearts. (B) Injury-dependent expression of lepb depicted by UMAP plot. (C) Enrichment of cells expressing the Cluster 10 specific genes (TFs), including lepb, cxcl8b.1, c3a.3 and steap4 |
|
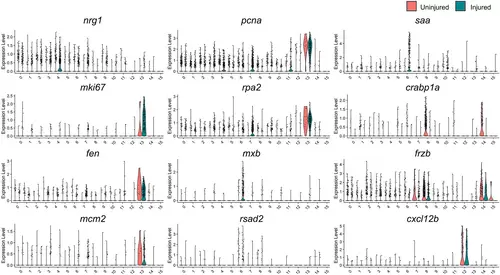
Epicardial gene expression in tcf21+ Epi/EPDCs of uninjured and regenerating hearts. Violin plot comparing the expression of nrg1, mki67, fen1, mcm2, PCNA, rpa2, mxb, rsad2, saa, crabp1a, Frzb, and cxcl12b to label distinct subpopulations of tcf21+ |
|
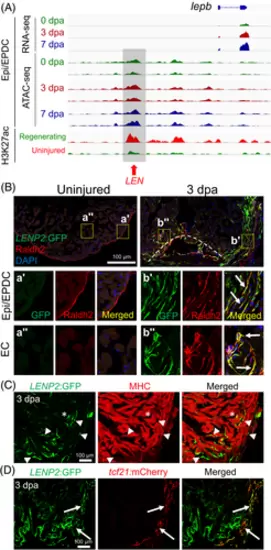
LEN directs injury-induced Epi/EPDC expression. (A) Genome browser tracks of the genomic region near lepb showing the transcripts and chromatin accessibility profiles in the Epi/EPDC. The whole-ventricle H3K27Ac profile of the uninjured and regenerating heart is shown at the bottom. Gray box and red arrow indicate LEN. (B-D) Immunostained section images of transgenic fish carrying LENP2:EGFP. (B) Raldh2 antibody is used to label EC and Epi/EPDC. Uninjured heart shows one single Raldh2+ cell layer outlining the cardiac chamber. Raldh2 signal emerges in the ECs at the wound area and EPDCs in the cortical layers upon injury, which are co-labelled with LENP2:EGFP (Arrows). The boxed areas are enlarged at the bottom panels. (C) Myosin heavy chain (MHC) antibody is used to label CMs. (D) tcf21:mCherry is used for Epi/EPDC expression. While LENP2:EGFP rarely colocalizes with MHC+ CMs (Arrowheads), a subset of LENP2:EGFP co-localizes with tcf21:mCherry (Arrows). Note that asterix indicates CM expression as a basal expression of the P2 minimal promoter. At least five hearts for uninjured and injured samples were examined and all animals displayed a similar expression pattern |
|
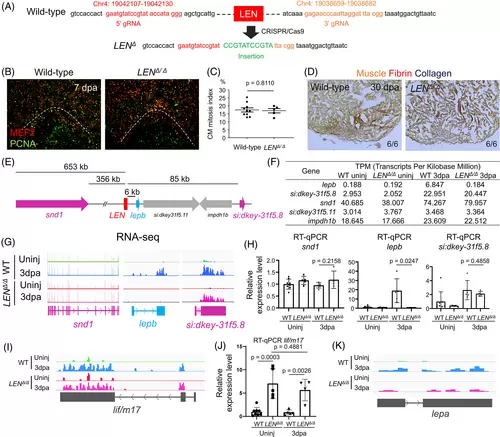
LEN is essential for injury-dependent lepb induction during heart regeneration. (A) Guide RNA sequences and genomic coordinates of LEN deletion (LENΔ) generated by CRISPR/Cas9. (B) Representative images of PCNA/Mef2 staining quantified in (C). (C) Quantification of adult ventricular CM proliferation indices for 7 dpa wild-type sibling (control) and LENΔ/Δ hearts. n = 10 and 5 for control and LENΔ/Δ hearts. Three sections per heart were used. (D) Representative images of AFOG staining for 30 dpa control and LENΔ/Δ hearts. n = 6 for control and LENΔ/Δ hearts. Three sections per heart were used. (E) Genomic region surrounding LEN. snd1, lepb, si:dkey-31f5.8 and rap1b are induced during heart regeneration. (F) Transcripts Per Kilobase Million (TPM) for lepb and its surrounding genes. (G) RNA-seq of uninjured and 3 dpa wild-type (WT) and LENΔ/Δ hearts. Tracks indicate an absence of lepb expression at 3 dpa in LENΔ/Δ fish, whereas snd1 and si:dkey-31f5.8 transcript levels are increased similarly in both wild type and LENΔ/Δ upon heart injury. (H) RT-qPCR analysis of snd1, lepb, and si:dkey-31f5.8 transcript levels in WT and LENΔ/Δ hearts. lepb is undetectable in 3 dpa injured LENΔ/Δ hearts. unpaired two-tailed t-tests were performed to indicate significance. (n = 8, 5, 5, and 4 for WT uninjured, LENΔ/Δ uninjured, WT 3dpa, and LENΔ/Δ 3dpa, respectively). Data are mean ± s.d. (I, J) lif/m17 is upregulated upon injury in wild-type and LENΔ/Δ hearts. (I) RNA-seq of uninjured and 3 dpa wild-type (WT) and LENΔ/Δ hearts. Tracks indicate lif/m17 transcript level is increased similarly in both wild type and LENΔ/Δ upon heart injury. (J) RT-qPCR analysis of lif/m17transcript level in WT and LENΔ/Δ hearts. Data are mean ± s.d. (K) RNA-seq of uninjured and 3 dpa wild-type (WT) and LENΔ/Δ hearts. Tracks indicate lepa transcript level is increased similarly in both wild type and LENΔ/Δ upon heart injury |