- Title
-
Profiling subcellular localization of nuclear-encoded mitochondrial gene products in zebrafish
- Authors
- Uszczynska-Ratajczak, B., Sugunan, S., Kwiatkowska, M., Migdal, M., Carbonell-Sala, S., Sokol, A., Winata, C.L., Chacinska, A.
- Source
- Full text @ Life Sci Alliance
|
|
|
|
|
|
|
|
|
|
|
|
|
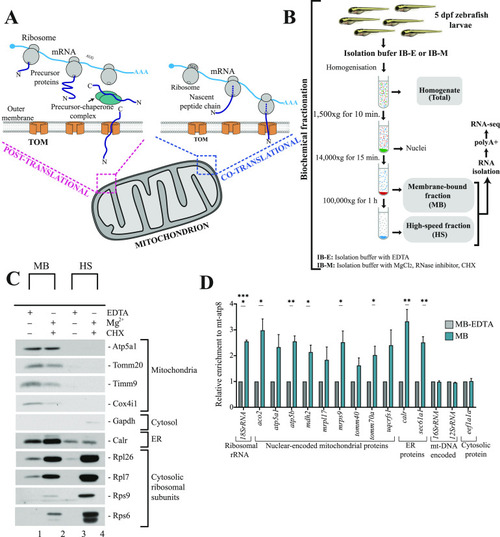
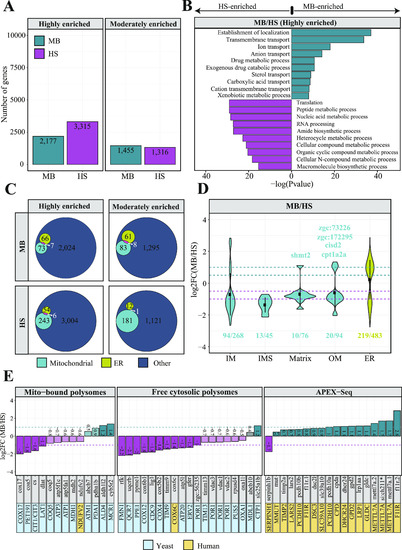
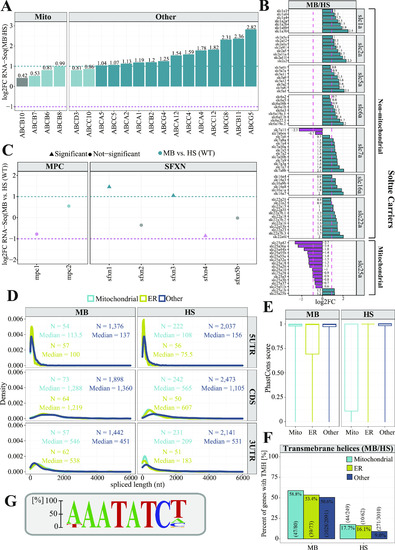
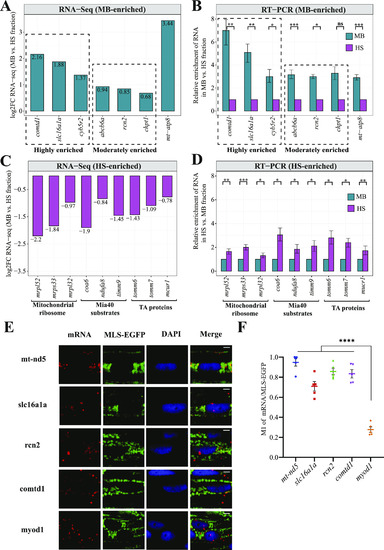
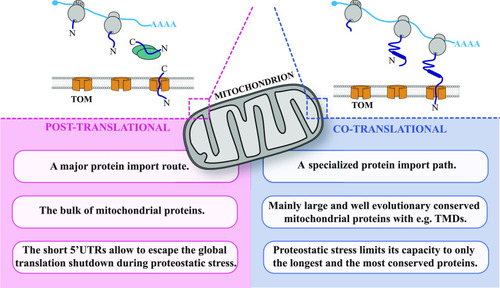
A model describing principles of mitochondrial protein import in zebrafish. |