- Title
-
A zebrafish model of congenital nephrotic syndrome of the Finnish type
- Authors
- Lee, M.S., Devi, S., He, J.C., Zhou, W.
- Source
- Full text @ Front Cell Dev Biol
|
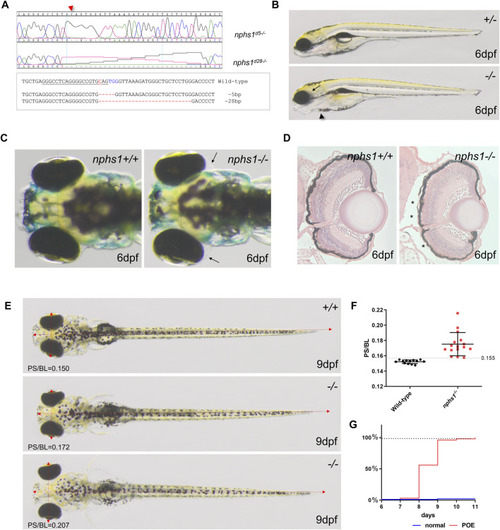
Generation of nphs1 mutant zebrafish by CRISPR/Cas9-mediated gene editing. |
|
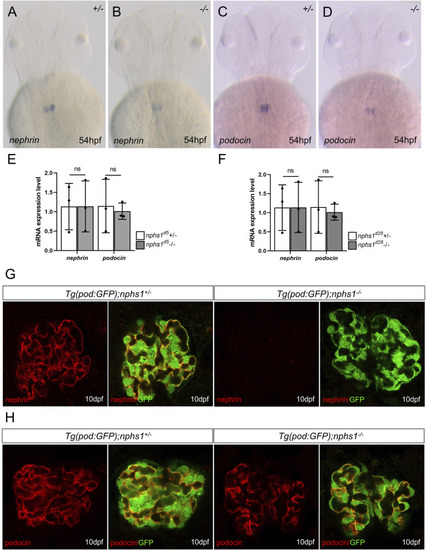
The expression analyses of nephrin and podocin in nphs1 mutants. Whole-mount |
|
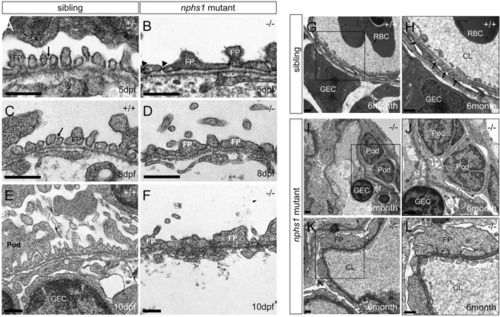
Lack of nephrin results in absence of slit-diaphragms and foot process effacement. |
|
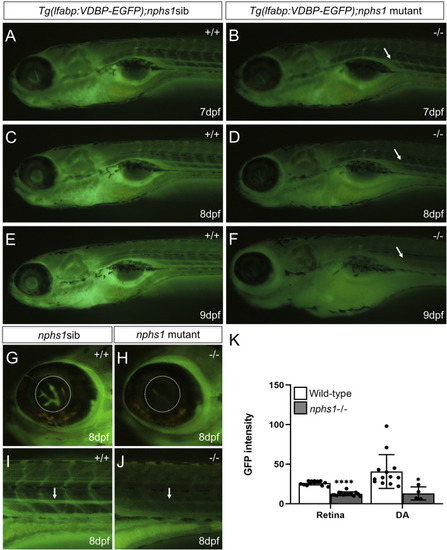
Measurement of hypoalbuminemia-like phenotype in nphs1-/- zebrafish. |