- Title
-
Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins
- Authors
- Osorio-Méndez, D., Miller, A., Begeman, I.J., Kurth, A., Hagle, R., Rolph, D., Dickson, A.L., Chen, C.H., Halloran, M., Poss, K.D., Kang, J.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
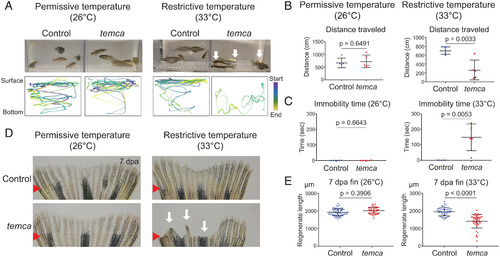
PHENOTYPE:
|
|
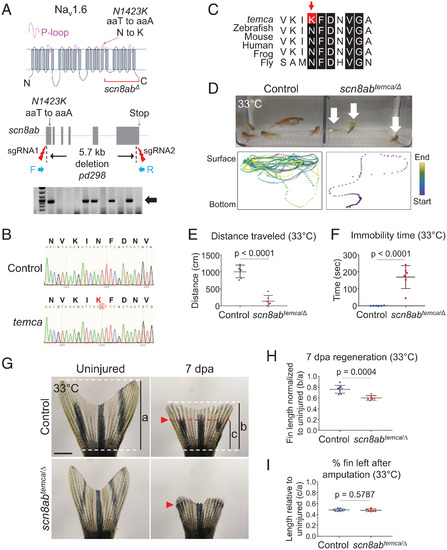
PHENOTYPE:
|
|
|
|
|
|
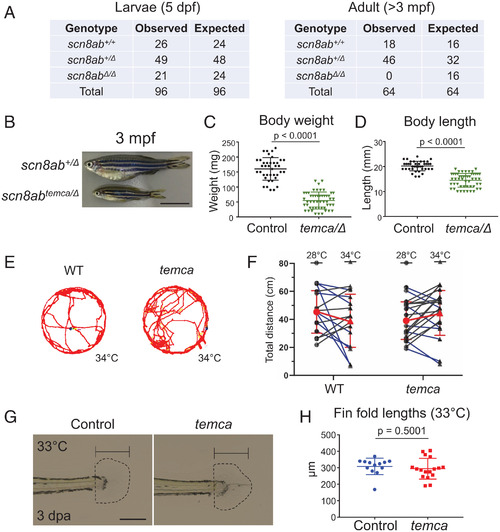
PHENOTYPE:
|
|
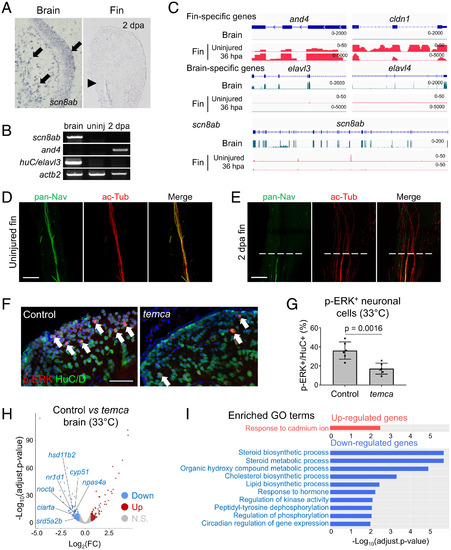
PHENOTYPE:
|