- Title
-
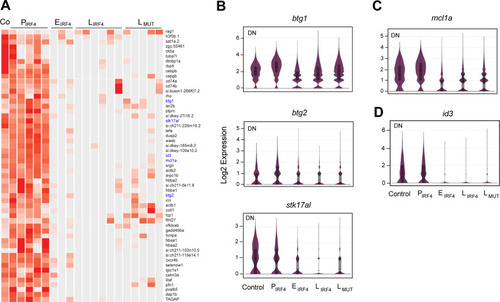
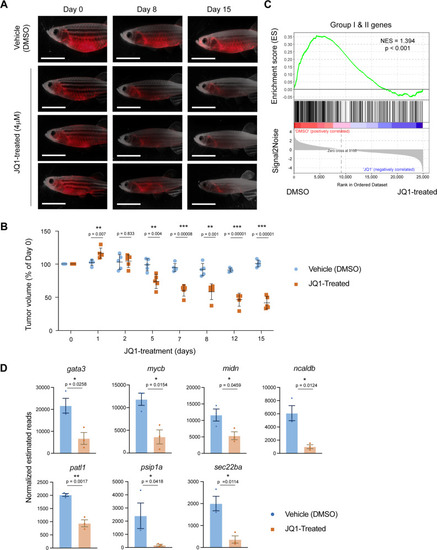
IRF4 drives clonal evolution and lineage choice in a zebrafish model of T-cell lymphoma
- Authors
- Amanda, S., Tan, T.K., Ong, J.Z.L., Theardy, M.S., Wong, R.W.J., Huang, X.Z., Ali, M.Z., Li, Y., Gong, Z., Inagaki, H., Foo, E.Y., Pang, B., Tan, S.Y., Iida, S., Sanda, T.
- Source
- Full text @ Nat. Commun.
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
|
|
|
|
|
|
|
|
|