- Title
-
Zebrafish imaging reveals TP53 mutation switching oncogene-induced senescence from suppressor to driver in primary tumorigenesis
- Authors
- Haraoka, Y., Akieda, Y., Nagai, Y., Mogi, C., Ishitani, T.
- Source
- Full text @ Nat. Commun.
|
|
|
|
|
PHENOTYPE:
|
|
|
|
|
|
|
|
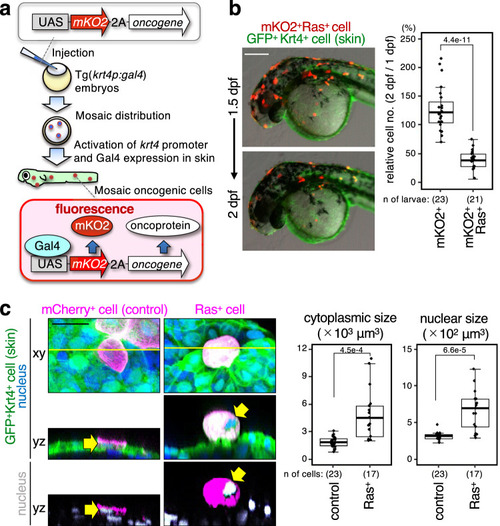
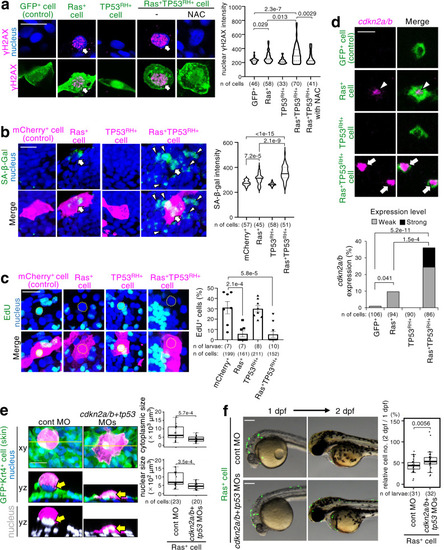
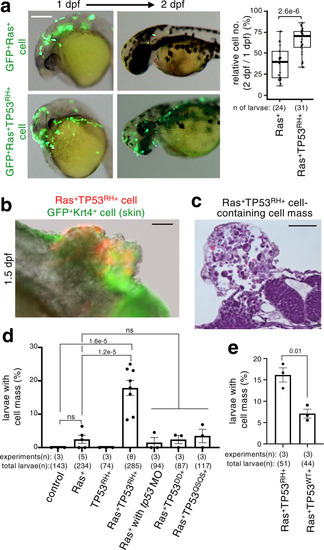
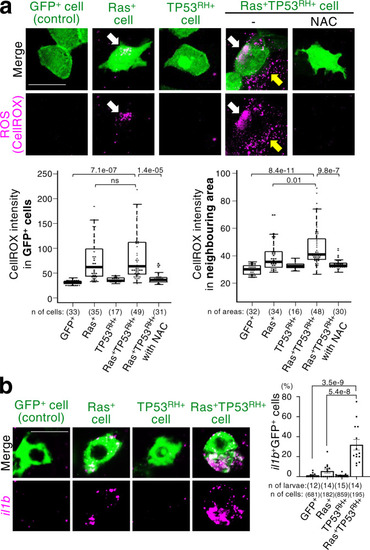
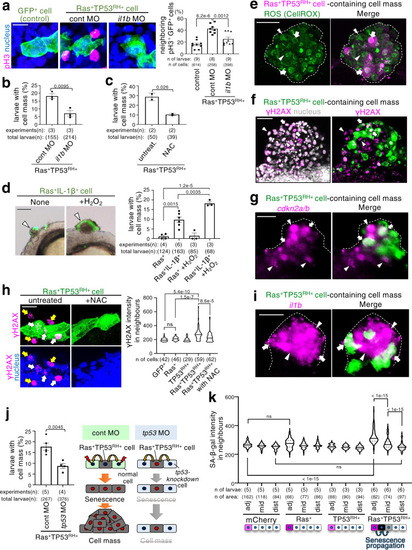
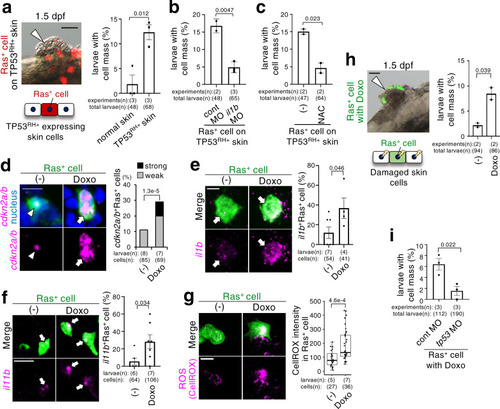
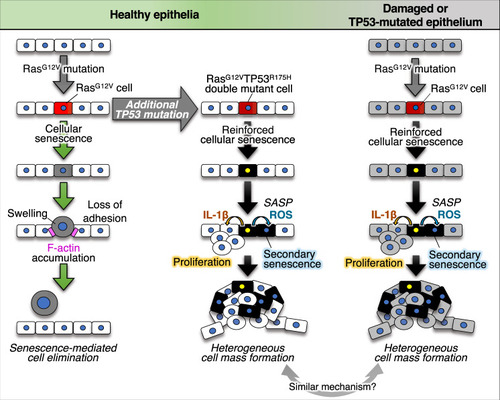
A newly emerged oncogenic cell becomes senescent and is eliminated from the epithelia. Additional TP53 mutation and tissue damage switches oncogene-induced senescence from suppressor to driver during primary tumorigenesis. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|