- Title
-
V3 Interneurons are Active and Recruit Spinal Motor Neurons During In Vivo Fictive Swimming in Larval Zebrafish
- Authors
- Wiggin, T.D., Montgomery, J.E., Brunick, A.J., Peck, J.H., Masino, M.A.
- Source
- Full text @ eNeuro
|
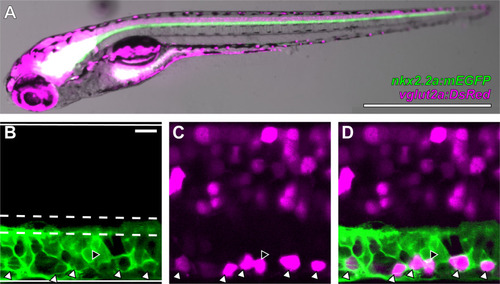
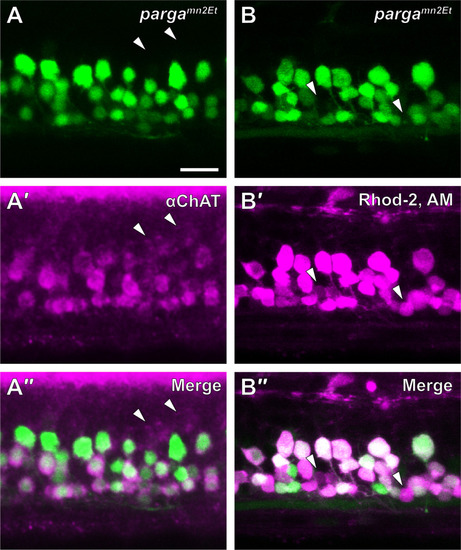
V3-INs are identified by co-localization of |
|
Distribution of V3-IN cell bodies in the spinal cord. |
|
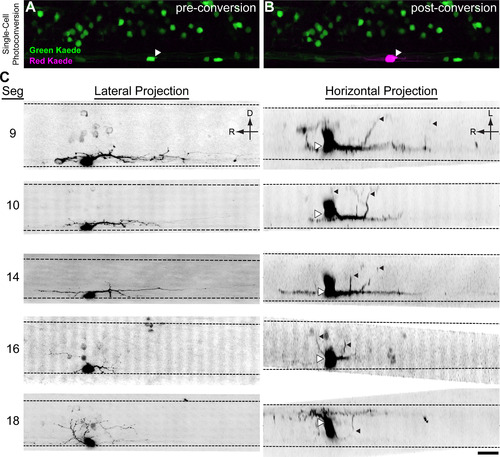
V3-INs are descending, bilaterally projecting cells. Kaede was expressed in |
|
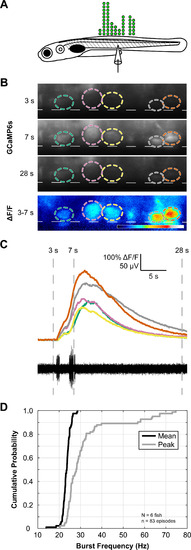
V3-IN activity is temporally correlated to fictive locomotion. |
|
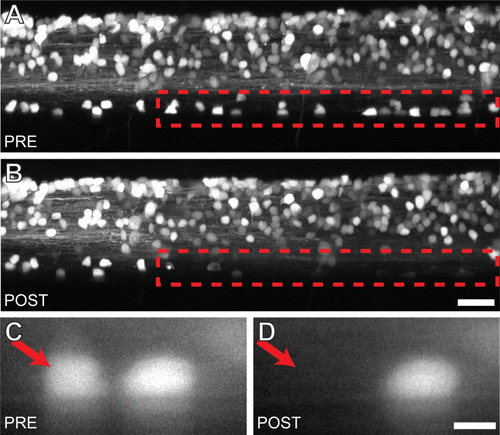
Targeted ablation of V3-INs is both efficient and spatially restricted. |
|
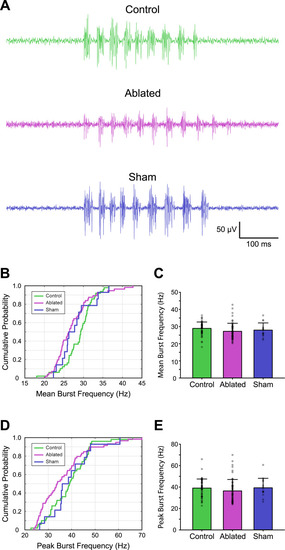
Targeted ablation of V3-INs does not affect burst frequency during spontaneous fictive locomotion. |
|
Spinal neurons loaded with rhodamine-2 AM dye are definitively identified as MNs. |
|
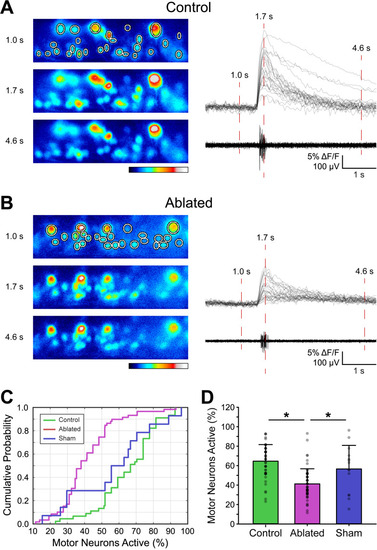
V3-IN laser ablation reduces the proportion of MNs that are active during fictive swimming. MNs in |