- Title
-
Cell Type-Specific Transcriptome Profiling Reveals a Role for Thioredoxin During Tumor Initiation
- Authors
- Korte, B.G., Giese, M.A., Ramakrishnan, G., Ma, S., Bennin, D., Rindy, J., Dewey, C.N., Huttenlocher, A.
- Source
- Full text @ Front Immunol
|
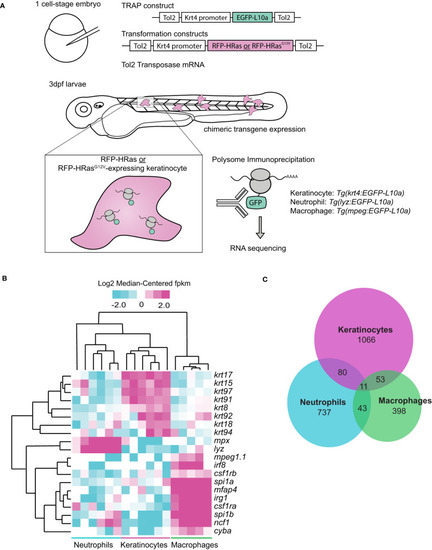
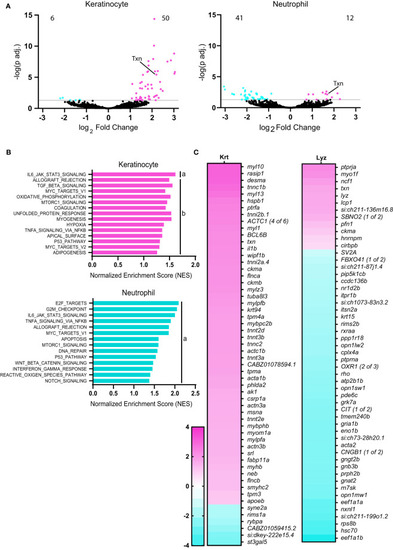
Oncogenic HRas induces differentially expressed genes in neutrophils and transformed cells in zebrafish larvae. |
|
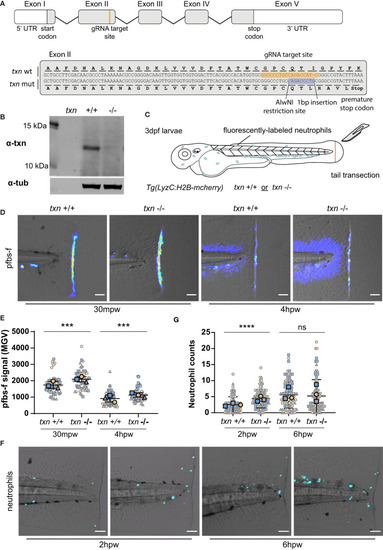
Thioredoxin expression is induced in neutrophils and keratinocytes with HRasG12V transformation. |
|
Thioredoxin regulates neutrophil recruitment and redox balance in damaged tissues. EXPRESSION / LABELING:
PHENOTYPE:
|
|
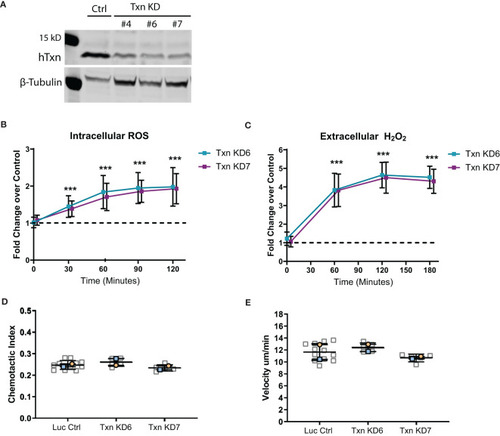
Thioredoxin depletion in a neutrophil-like cell line increases ROS but does not intrinsically affect motility. |
|
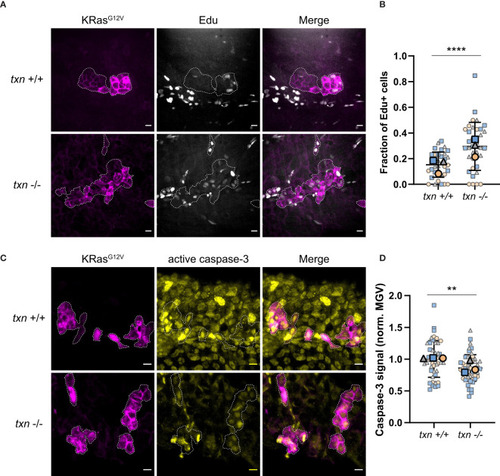
Transformed keratinocytes exhibit increased proliferation and reduced apoptosis during tumor initiation in |
|
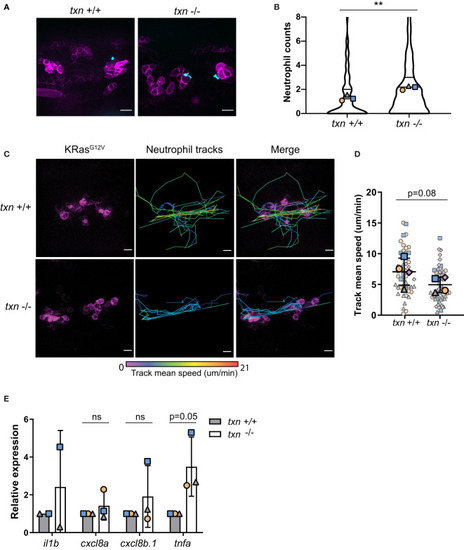
Thioredoxin affects neutrophil motility around KRasG12V-transformed keratinocytes. |
|
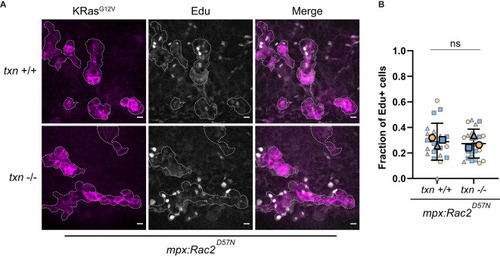
Transformed keratinocyte proliferation is not altered in |