- Title
-
Histone 3 Methyltransferases Alter Melanoma Initiation and Progression Through Discrete Mechanisms
- Authors
- DiNapoli, S.E., Martinez-McFaline, R., Shen, H., Doane, A.S., Perez, A.R., Verma, A., Simon, A., Nelson, I., Balgobin, C.A., Bourque, C.T., Yao, J., Raman, R., Béguelin, W., Zippin, J.H., Elemento, O., Melnick, A.M., Houvras, Y.
- Source
- Full text @ Front Cell Dev Biol
|
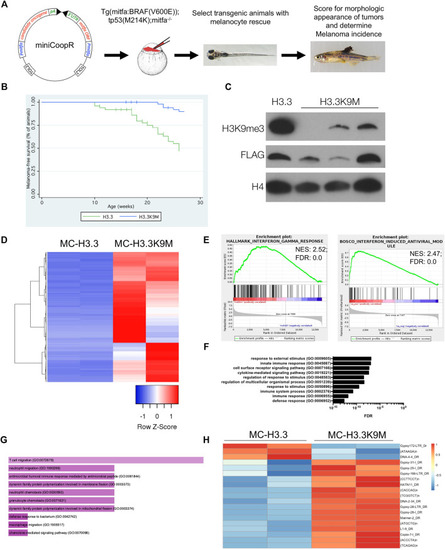
H3.3K9M suppresses melanoma formation. |
|
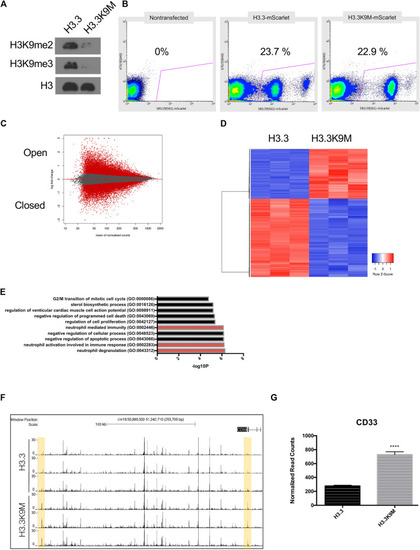
H3.3K9M induces interferon response. |
|
A-366 induces inflammatory response with no effect on cellular growth. |
|
PRC2 alterations in melanoma. |
|
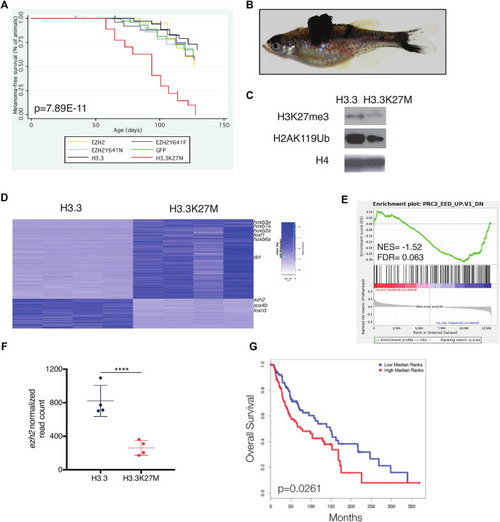
H3.3K27M accelerates melanoma formation. |
|
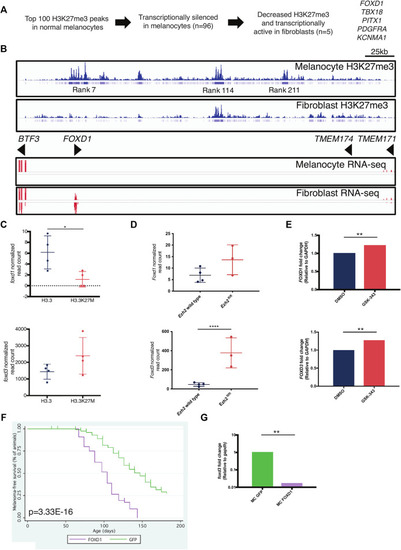
FOXD1 is a PRC2 target gene in melanocytes. |