- Title
-
A novel deep learning-based 3D cell segmentation framework for future image-based disease detection
- Authors
- Wang, A., Zhang, Q., Han, Y., Megason, S., Hormoz, S., Mosaliganti, K.R., Lam, J.C.K., Li, V.O.K.
- Source
- Full text @ Sci. Rep.
|
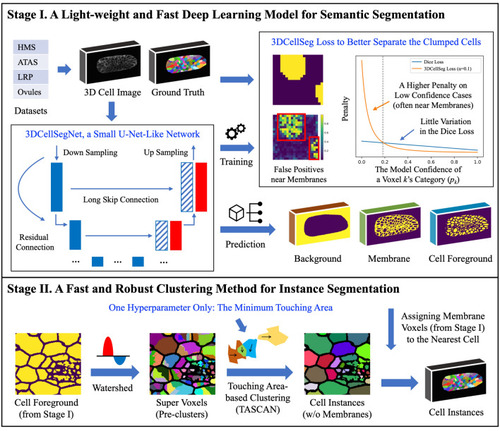
3DCellSeg: A two-stage light-weight, fast, and robust pipeline for 3D cell segmentation. [Note: There are two stages in the pipeline. The first stage is a semantic segmentation, where the input is a 3D cell membrane image and the output consists of three masks, which indicate whether a voxel is the cell foreground, membrane, or background. The second stage is an instance segmentation performed on the basis of these three masks. The cellular images and segmentation results were generated by Python Matplotlib ( |
|
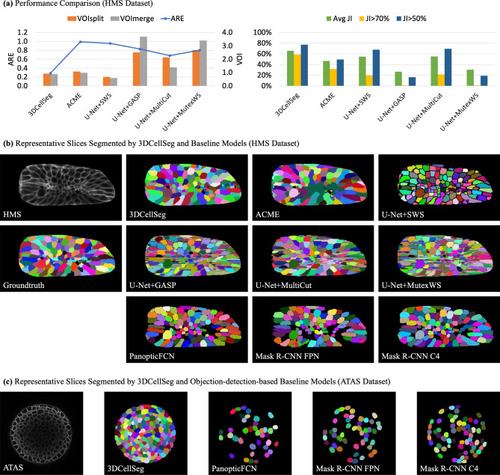
Model comparison and representative slices. [ |
|
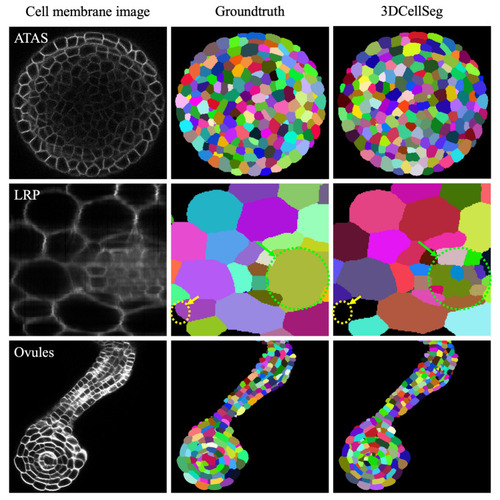
3DCellSeg performance on the ATAS, LRP, and Ovules datasets. [Note: Different cell instances were randomly assigned different colors. The LRP dataset images are annotated: the yellow circle shows where 3DCellSeg has made a mistake and the green circle shows that 3DCellSeg can segment cells that were not labelled in the ground truth. The cellular images were generated by Python Matplotlib ( |
|
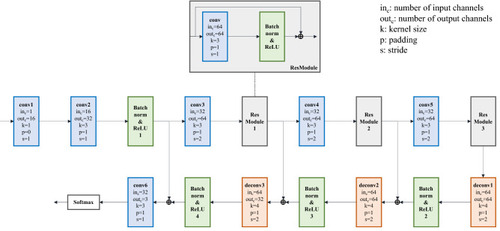
The structure of 3DCellSegNet. [Note: The extra voxels on the edge of the feature maps are removed after each deconvolution operation, in order to ensure the size of the up-sampled feature map is identical with that of the corresponding down-sampled feature map]. |
|
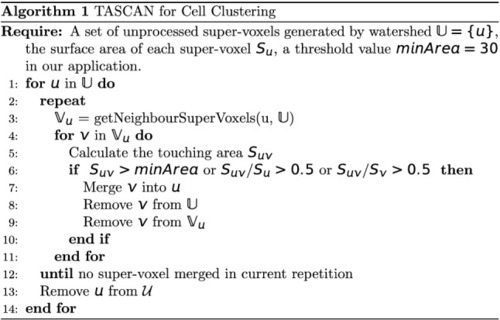
TASCAN algorithm for cell clustering. |