- Title
-
Zebrafish mbnl mutants model physical and molecular phenotypes of myotonic dystrophy
- Authors
- Hinman, M.N., Richardson, J.I., Sockol, R.A., Aronson, E.D., Stednitz, S.J., Murray, K.N., Berglund, J.A., Guillemin, K.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
|
|
PHENOTYPE:
|
|
|
|
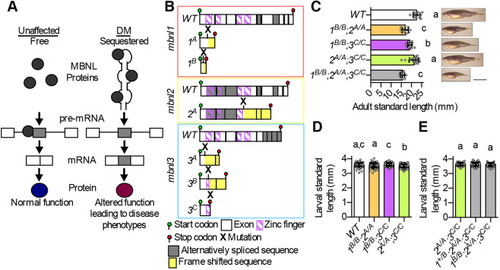
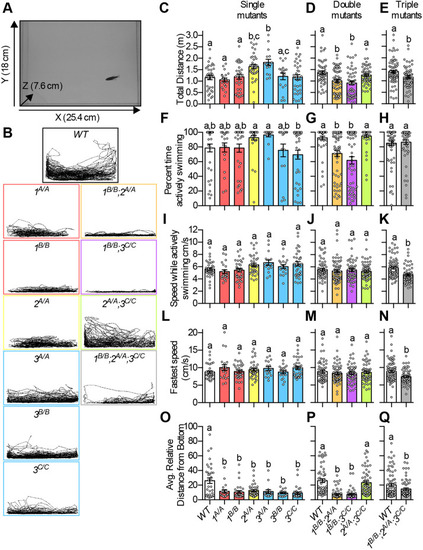
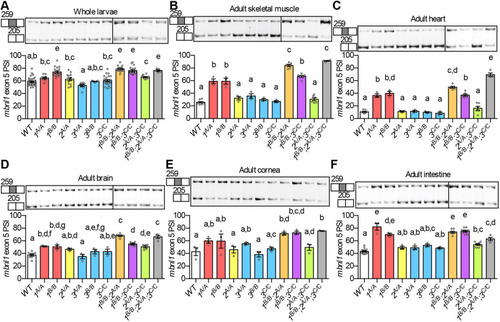
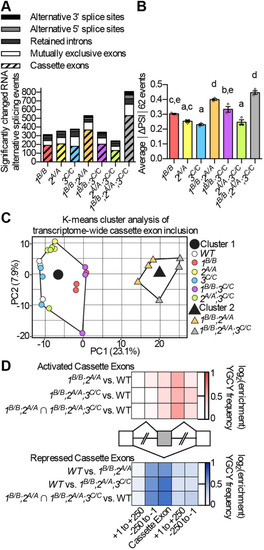
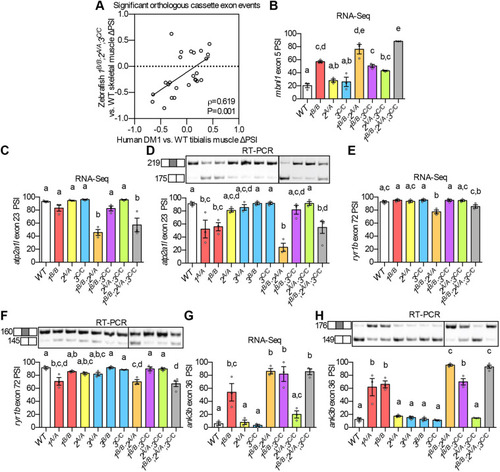
(A) ΔPSI between mutant and WT are shown for orthologous exons in zebrafish 1B/B;2,sup>A/A;3C/C skeletal muscle and in tibialis muscle from human DM1 patients. (B-H) RNA-Seq (B,C,E,G) and RT-PCR (D,F,H) analyses showing PSI of mbnl1 exon 5 (B), atp2a1l exon 23 (C,D), ryr1b exon 72 (E,F), and ank3b exon 36 (G,H) in WT and mbnl mutant adult zebrafish skeletal muscle. Orthologous human exon inclusion in DM patients is increased for MBNL1 and ANK3, and decreased for ATP2A1 and RYR1 (Freyermuth et al., 2016; Gates et al., 2011; Kimura et al., 2005; Terenzi and Ladd, 2010; Tran et al., 2011; Zhao et al., 2015). In A-H, mbnl1 mutant alleles are denoted as 1A and 1B, mbnl2 alleles as 2A, and mbnl3 alleles as 3A, 3B and 3C. In A, ρ is the Spearman's rank correlation coefficient. In B-H, data are presented as mean±s.e.m. Each dot represents RNA from one fish. Representative gels are shown above each RT-PCR graph with band sizes in bp shown on the left. White boxes represent constitutive exons and gray boxes represent alternative exons. Dividing lines indicate samples run on separate gels. Data were analyzed by ordinary one-way ANOVA with Tukey's multiple comparisons test. Data bars that do not share the same letter above them are significantly different from one another. Raw data and statistical analysis details are in Table S5. PHENOTYPE:
|