- Title
-
Complementary and divergent functions of zebrafish Tango1 and Ctage5 in tissue development and homeostasis
- Authors
- Clark, E.M., Link, B.A.
- Source
- Full text @ Mol. Biol. Cell
|
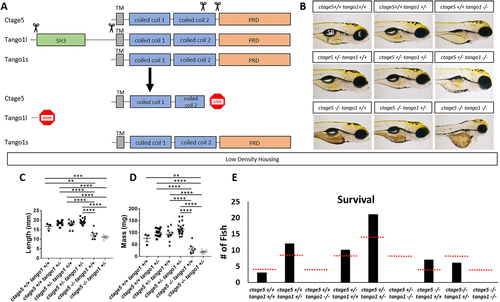
ctage5 and tango1 mutations affect size and survival of zebrafish: (A) Schematic showing CRISPR guide RNA cut sites (scissors) and the estimated final truncated proteins. (B) Representative brightfield images of all combinations of ctage5 and tango1 mutations in 4dpf zebrafish larvae. Y = yolk, SB = swim bladder, E = eye. (C) Length (one-way ANOVA, F = 38.6, p < 0.0001), (D) mass (one-way ANOVA, F = 28.22, p < 0.0001) and (E) survival (χ2 = 24.59, p < 0.05, n = 59) measurements of all combinations of ctage5 and tango1 mutant zebrafish at 2 mo raised in a low-density environment (about 10 fish per tank). Red-dotted lines in E represent expected survival, and black bars are actual survival. ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. PHENOTYPE:
|
|
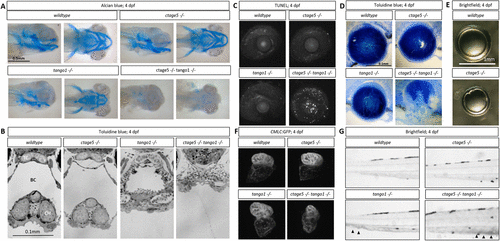
Craniofacial and other morphological defects in tango1 mutants are accentuated by ctage5 deletion. (A) Representative lateral (left) and ventral (right) images of Alcian blue–stained 4dpf larvae. (B) Representative images of plastic-sectioned and toluidine blue–stained 4dpf larvae cropped to show craniofacial alterations. TC = trabeculae, BC = buccal cavity, CH = ceratohyal cartilage. (C) Representative maximum intensity projection images showing TUNEL staining of 4dpf lens and sclera. (D) Representative images of plastic-sectioned and toluidine blue–stained 4dpf lens. (E) Representative images of lens dissected from 3-mo-old zebrafish. (F) Representative images showing ventricle-enriched cardiac myosin light chain (CMLC:GFP) expression. (G) Representative brightfield image of the trunk for each genotype. Arrowheads detail blood pooling in the caudal vein. PHENOTYPE:
|
|
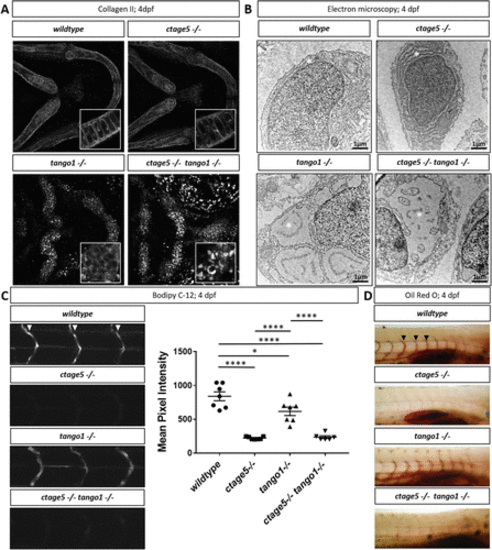
Trafficking differences in ctage5 vs. tango1 mutant embryos. (A) Representative 40× images showing collagen 2 wholemount immunohistochemistry in 4dpf larvae chondrocytes (n = 3–4 per group). Inserts are 40× images with 3× zoom and cropped to show higher-resolution chondrocyte collagen 2 staining. (B) Representative electron microscopy images of endoplasmic reticulum (*) in 4dpf larvae chondrocytes (n = 3 per group). (C) Representative images and quantification of BODIPY C-12 fluorescence in the intersegmental vessels (arrowheads; one-way ANOVA, F = 39.36, p < 0.0001). (D) Representative images of Oil Red O staining for lipids in 4dpf zebrafish larvae intersegmental vessels (arrowheads). * = p < 0.05, **** = p < 0.0001. |
|
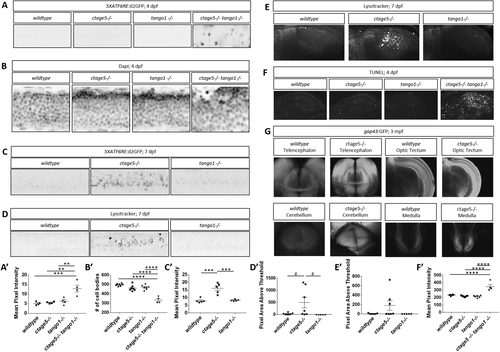
Stress pathway activation in the nervous system is specific for ctage5 -/- and amplified by tango mutation. (A, A’) Representative images, A, and quantification, A’, of 5XATF6RE:d2GFP activation in 4dpf spinal cords (one-way ANOVA, F = 12.42, p = 0.0005). (B, B’) Representative images of DAPI staining, B, and quantification of brightfield images, B’, in 4dpf spinal cords to show cellular organization (asterisk shows cells mislocalized from the normal lamination pattern; one-way ANOVA, F = 37.51, p < 0.0001). (C, C’) Representative images, C, and quantification, C’, of 5XATF6RE:d2GFP activation in 7dpf spinal cords (one-way ANOVA, F = 22.22, p < 0.0001). (D, D’) Representative images, D, and quantification, D’, of the lysotracker staining area above threshold for acidic compartments in 7dpf spinal cords (one-way ANOVA, F = 3.881, p = 0.0423). (E, E’) Representative images, E, and quantification, E’, of the lysotracker staining area above threshold of lysotracker for acidic compartments in 7dpf brains (one-way ANOVA, F = 2.479, p = 0.1154). (F, F’) Representative images, F, and quantification, F’, of TUNEL staining for cell death in 4dpf brains (one-way ANOVA, F = 24.33, p < 0.0001). (G) Representative images of gap43:GFP in 3–mo old PACT cleared ctage5 wildtype (WT) or homozygous mutant brains. # = p < 0.08, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. |
|
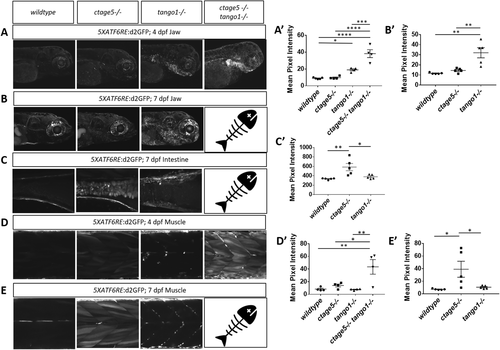
ER stress is prevalent in tissues outside of the nervous system. Representative images and quantification of 5XATF6RE:d2GFP ER stress in the jaw at 4dpf (A, A’; one-way ANOVA, F = 34.44, p < 0.0001) and 7dpf (B, B’; one-way ANOVA, F = 13.86, p < 0.0008), intestine at 7dpf (C, C’; one-way ANOVA, F = 8.358, p = 0.0053), and muscle at 4dpf (D, D’; one-way ANOVA, F = 8.205, p = 0.0031) and 7dpf (E, E’: F = 5.624, p = 0.0189). * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. Fish = indicates ctage5-/- tango1-/- mutants do not survive to 7dpf. |
|
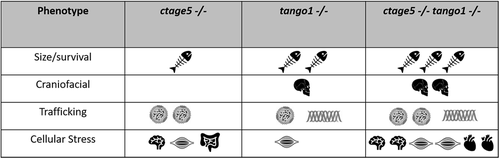
Summary of phenotype magnitude in mutant fish. Table summarizing phenotype severity. Any symbol represents an elevated severity compared with wild-type. More symbols represent increased severity. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

Unillustrated author statements PHENOTYPE:
|