- Title
-
Isthmin1, a secreted signaling protein, acts downstream of diverse embryonic patterning centers in development
- Authors
- Kesavan, G., Raible, F., Gupta, M., Machate, A., Yilmaz, D., Brand, M.
- Source
- Full text @ Cell Tissue Res.
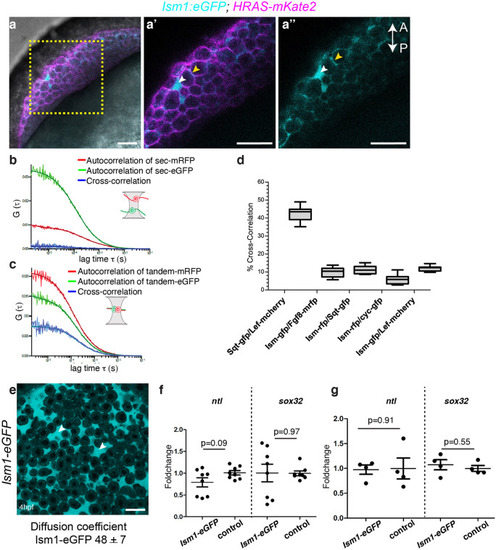
|
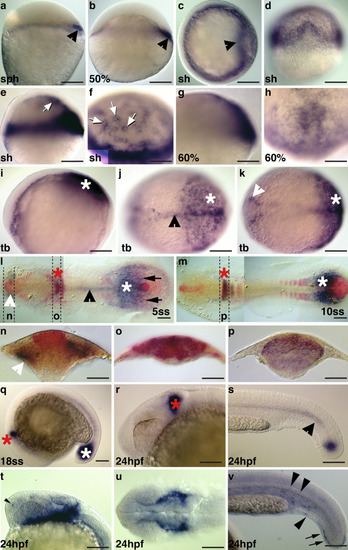
Expression of |
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
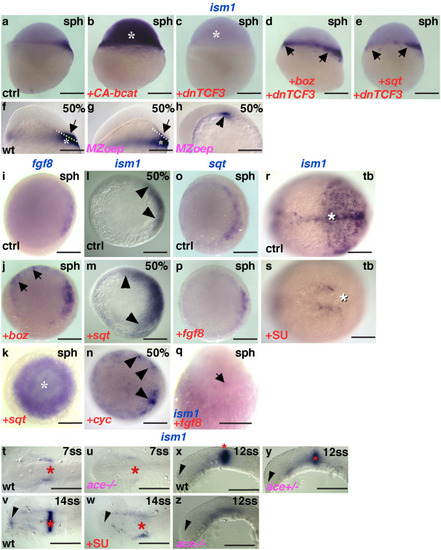
Dorsalization of embryos in EXPRESSION / LABELING:
|
|
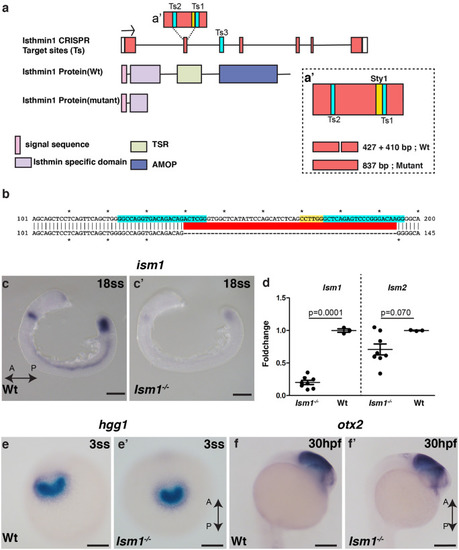
Generation and characterization of EXPRESSION / LABELING:
PHENOTYPE:
|
|
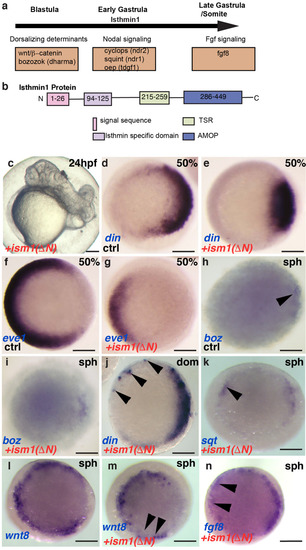
Ism1 interaction with Fgf8 and Nodal molecules in vivo |