- Title
-
eys+/- ; lrp5+/- Zebrafish Reveals Lrp5 Can Be the Receptor of Retinol in the Visual Cycle
- Authors
- Takita, S., Seko, Y.
- Source
- Full text @ iScience
|
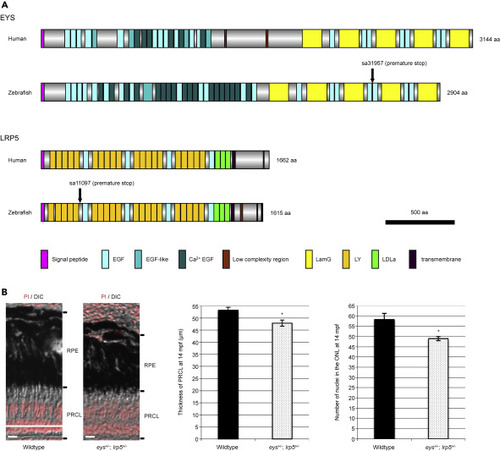
Figure 1. Conservation of Human and Zebrafish EYS and LRP5 Proteins and Mild Photoreceptor Degeneration in the Digenic eys+/-; lrp5+/− Zebrafish (A) The domain structures of human and zebrafish EYS and LRP5 proteins show that the compositions of EYS and LRP5 proteins are well conserved between the two species. Human and zebrafish EYS proteins consist of a signal peptide sequence for secretion, epidermal growth factor (EGF), EGF-like and Ca2+ EGF domains in the N-terminal half, and laminin A G-like (LamG) and EGF domains in the C-terminal half. Human and zebrafish LRP5 proteins consist of a signal peptide sequence, low-density lipoprotein receptor YWTD (LY; also known as β-propeller), EGF, and LDLa domains in the N-terminal extracellular half followed by transmembrane region and low complexity region in the C-terminal intracellular region. Vertical arrows in zebrafish EYS and LRP5 proteins indicate the point mutations resulting in premature stop codons (nonsense mutations). (B) The thickness of photoreceptor cell layer (PRCL) was 10% reduced in the eys+/-; lrp5+/− zebrafish eyes compared with wild-type PRCL at 14 mpf (wild-type, 53.3 ± 1.1 μm; eys+/-; lrp5+/− mutants, 47.9 ± 1.2 μm). The number of nuclei in the ONL was reduced 16% in the eys+/-; lrp5+/− zebrafish eyes compared with wild-type PRCL (wild-type, 58.3 ± 3.1; eys+/-; lrp5+/− mutants, 49 ± 1.0). Data are represented as mean ± SD. ∗p < 0.01. DIC, differential interference contrast; mpf, months post fertilization; RPE, retinal pigment epithelium. Scale bars, 10 μm. PHENOTYPE:
|
|
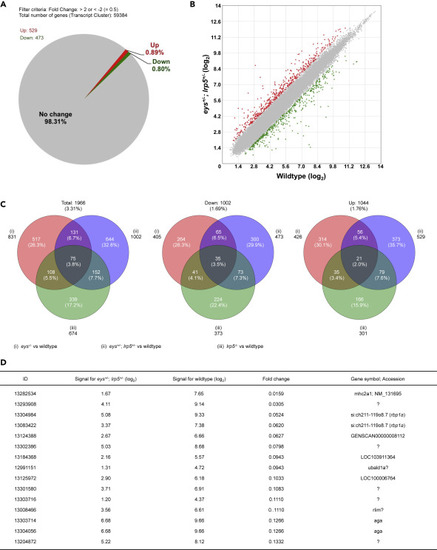
Figure 2. Global Gene Expression Analysis of the Digenic eys+/-; lrp5+/− Zebrafish Eyes Identifies the retinol binding protein 1a (rbp1a) Gene. (A and B) About 1.69% (1,002 genes) were 2-fold changed among the 59,384 transcript clusters (genes) in the eys+/-; lrp5+/− zebrafish eyes compared with the eyes of wild-type siblings (A). About 0.80% (473 genes) of the total were 2-fold downregulated and 0.89% (529 genes) were 2-fold upregulated (B, green and red spots, respectively). (C) About 3.31% (1,966 genes) were 2-fold changed, 1.69% (1,002) genes were 2-fold downregulated, and 1.76% (1,044 genes) were 2-fold upregulated in the eys−/−, lrp5−/−, or eys+/-; lrp5+/− eyes compared with the eyes of wild-type siblings (left, middle, and right panels, respectively). Among 0.80% (473 genes) 2-fold downregulated in the eys+/-; lrp5+/− eyes (middle panel), 21.1% (100 genes) were overlapped with the eyes of eys−/− siblings and 14.6% (108 genes) were overlapped with the eyes of lrp5−/− siblings. (D) Genes whose expression levels were dramatically changed were extracted. “Fold change” in the fourth column indicates the ratio (the bare value of the signal for eys+/-; lrp5+/− divided by that for the wild-type). The ratio for si:ch211-119o8.7 (the rbp1a gene, third and fourth rows) was identified as the gene remarkably downregulated in the eys+/-; lrp5+/− eyes (fold changes, 0.0524 (ID: 13304984) and 0.0620 (13083422)). “Signal for eys+/-; lrp5+/−” (second column) and “Signal for wild type” (third column) are shown in (log2). |
|
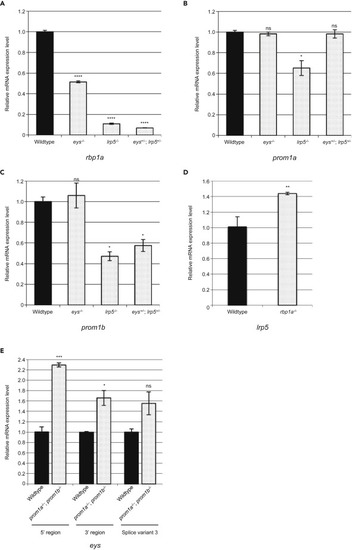
Figure 3. The rbp1a Gene Plays a Role Downstream of the lrp5 Gene (A) qPCR analysis showed that the expression level of the rbp1a gene was remarkably decreased in the lrp5−/− (~11%) and digenic eys+/-; lrp5+/− (~7%) eyes while moderately decreased in the eys−/− eyes (~51%) compared with wild-type eyes at 3.5 mpf. (B and C) There was no change in the expression level of either the prom1a or prom1b gene in the eys−/− eyes compared with wild-type eyes at 3.5 mpf. (D) The expression level of the lrp5 gene was not remarkably changed in the rbp1a−/− eyes (~142%) compared with wild-type eyes at 3.5 mpf. The rbp1a gene played a role downstream of the lrp5 gene (A and D). (E) The expression level of the eys gene was slightly increased in the prom1a−/−; prom1b−/− eyes (~229% for 5′ region, ~166% for 3′ region, and ~155% for splice variant 3 [Takita et al., 2019]) compared with wild-type eyes at 3.5 mpf. Negative genetic interaction between the eys gene and the prom1a or prom1b gene was not observed (B, C, and E). All data are represented as mean ± SD. ∗p < 0.005, ∗∗p < 0.0005, ∗∗∗p < 0.0001, ∗∗∗∗p < 0.000001. Ns, not significant. |
|
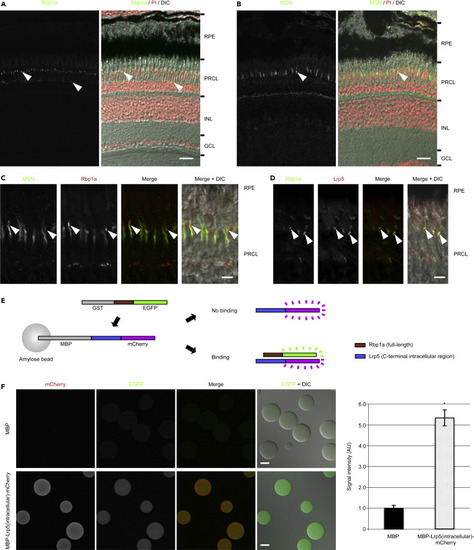
Figure 4. Lrp5 and Rbp1a Proteins Are Colocalized in the Outer Retinas and Bind Directly In Vitro. (A) Staining of Rbp1a protein was observed in the outer retina (arrowheads). Nuclei were stained with propidium iodide (PI) (red). (B and C) Rbp1a signal overlapped well with MOESIN (MSN), a marker of the microvilli of the RPE, and strong signal was observed at the tip (arrowheads), which suggested that Rbp1a was concentrated in the tip of the microvilli in the RPE. (D) Staining of Lrp5 protein with Rbp1a protein revealed that Lrp5 was colocalized with Rbp1a at the concentrated region (tip) of the microvilli of the RPE (arrowheads). (E) Experimental design of binding assay. Binding is detected by fluorescence from EGFP. (F) EGFP signal was observed on the beads in which the intracellular region of Lrp5 protein was fused to MBP (lower panels), whereas it was not detected on the beads in which MBP alone was present (upper panels) (MBP, 1.0 ± 0.1 for 8 beads; MBP-Lrp5(intracellular)-mCherry, 5.3 ± 0.4 for 6 beads). Data are represented as mean ± SD. ∗p < 0.0000005. DIC, differential interference contrast; GCL, ganglion cell layer; INL, inner nuclear layer; PRCL, photoreceptor cell layer; RPE, retinal pigment epithelium. Scale bars, 20 μm in (A) and (B), 5 μm in (C) and (D), and 50 μm in (F). EXPRESSION / LABELING:
|
|
Figure 5. A Proposed Model of LRP5 Function in the Visual Cycle LRP5 protein (shown in orange in right bottom) is a strong candidate for the receptor of atROL by forming a complex with RBP1 protein (the transporter of atROL, shown in light green in right bottom) and may be involved in the uptake of atROL in the RPE in the visual cycle. The flow of retinoids is shown by red arrows and the unknown possible interactions are indicated with “?”. atROL, all-trans retinol; ABCA4, ATP binding cassette subfamily A member 4; IRBP, inter-photoreceptor retinol binding protein (also known as RBP3, retinol binding protein 3); LRAT, lecithin retinol acyltransferase; LRP5, LDL receptor related protein 5; RBP1, retinol binding protein 1 (also known as CRBP, cellular retinol binding protein); RDH, retinol dehydrogenase; RLBP1, retinaldehyde binding protein 1 (also known as CRALBP, cellular retinal binding protein); RPE65, retinoid isomerohydrolase RPE65; STRA6, stimulated by retinoic acid gene 6. |