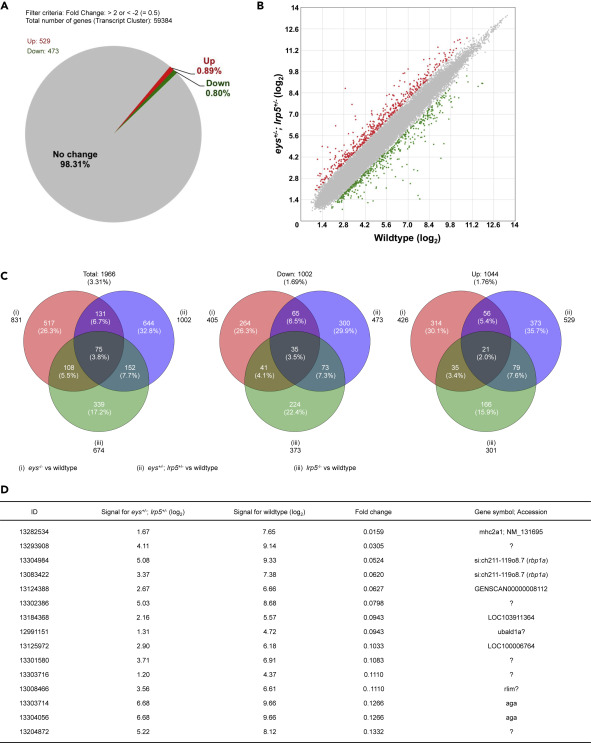
Fig. 2 Figure 2. Global Gene Expression Analysis of the Digenic eys+/-; lrp5+/− Zebrafish Eyes Identifies the retinol binding protein 1a (rbp1a) Gene. (A and B) About 1.69% (1,002 genes) were 2-fold changed among the 59,384 transcript clusters (genes) in the eys+/-; lrp5+/− zebrafish eyes compared with the eyes of wild-type siblings (A). About 0.80% (473 genes) of the total were 2-fold downregulated and 0.89% (529 genes) were 2-fold upregulated (B, green and red spots, respectively). (C) About 3.31% (1,966 genes) were 2-fold changed, 1.69% (1,002) genes were 2-fold downregulated, and 1.76% (1,044 genes) were 2-fold upregulated in the eys−/−, lrp5−/−, or eys+/-; lrp5+/− eyes compared with the eyes of wild-type siblings (left, middle, and right panels, respectively). Among 0.80% (473 genes) 2-fold downregulated in the eys+/-; lrp5+/− eyes (middle panel), 21.1% (100 genes) were overlapped with the eyes of eys−/− siblings and 14.6% (108 genes) were overlapped with the eyes of lrp5−/− siblings. (D) Genes whose expression levels were dramatically changed were extracted. “Fold change” in the fourth column indicates the ratio (the bare value of the signal for eys+/-; lrp5+/− divided by that for the wild-type). The ratio for si:ch211-119o8.7 (the rbp1a gene, third and fourth rows) was identified as the gene remarkably downregulated in the eys+/-; lrp5+/− eyes (fold changes, 0.0524 (ID: 13304984) and 0.0620 (13083422)). “Signal for eys+/-; lrp5+/−” (second column) and “Signal for wild type” (third column) are shown in (log2).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ iScience