- Title
-
ALX1-related frontonasal dysplasia results from defective neural crest cell development and migration
- Authors
- Pini, J., Kueper, J., Hu, Y.D., Kawasaki, K., Yeung, P., Tsimbal, C., Yoon, B., Carmichael, N., Maas, R.L., Cotney, J., Grinblat, Y., Liao, E.C.
- Source
- Full text @ EMBO Mol. Med.
|
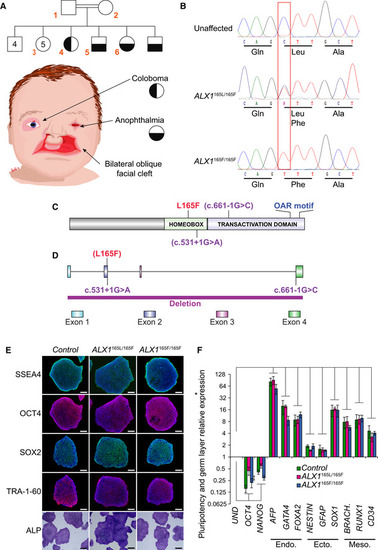
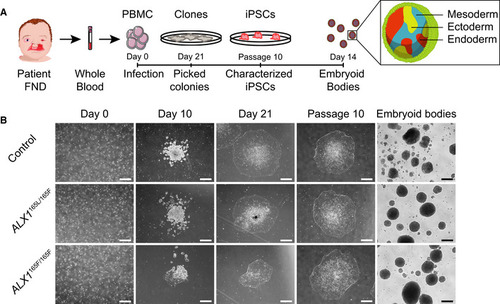
The pedigree family tree includes two unaffected parents, four unaffected male siblings, five unaffected female siblings, and two each female and male affected sibling. Subjects 1–6, indicated in red, were enrolled in the study. Subjects 4–6 show complex FND with ocular involvement. The eldest affected sibling (subject 4) presented with right coloboma, left microphthalmia, and bilateral Tessier 4 oblique facial clefts. Subject 5 presented with bilateral anophthalmia with fused eyelids and shallow orbits, with bilateral oblique facial clefts. Subject 6 presented with bilateral anophthalmia with open shallow orbits, absent upper and lower eyelids, exposed orbital mucosa, bilateral oblique facial clefts, and malformed nasal ala with nodular skin tags. iPSCs were generated using blood samples collected from subjects 1, 5, and 6. Whole‐exome sequencing was carried out and analysis revealed a missense p.L165F variant (c.493 C>T) in the Schematic of the ALX1 protein structure showing the position of the L165F substitution described here (red) and the locations of exon borders affected by two reported pathogenic variants (purple; Ullah Schematic of the Immunofluorescence staining for pluripotent markers Expression of pluripotent (OCT4, NANOG), endoderm (Endo., AFP, GATA4, FOXA2), ectoderm (Ecto., NESTIN, GFAP, SOX1), and mesoderm (Meso., BRACH. (BRACHYURY), RUNX1, CD34) gene markers for |
|
Schematic of the differentiation protocol timeline. Maintenance Medium (MM) = iPSC medium (StemFlex with 1× penicillin/streptomycin), NCC differentiation medium = DMEM‐F12, 10% fetal bovine serum, 1 mM sodium pyruvate, 1 mM penicillin/streptomycin, 1 mM nonessential amino acids, 110 μM 2‐mercaptoethanol, 10 ng/ml epidermal growth factor. Images of iPSC and iPSC‐derived NCC at Days 0, 14, and passage 4 following differentiation. Scale bars: 400 μm (Day 0), 200 μm (Day 14, passage 4). |
|
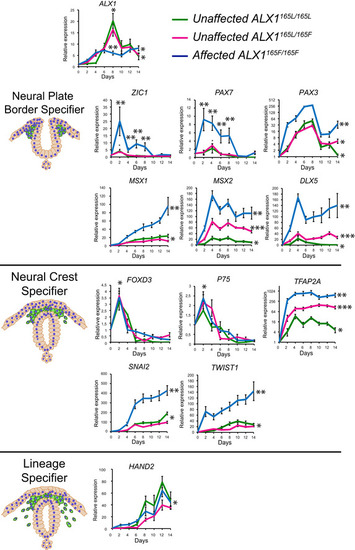
Gene expression analysis across |
|
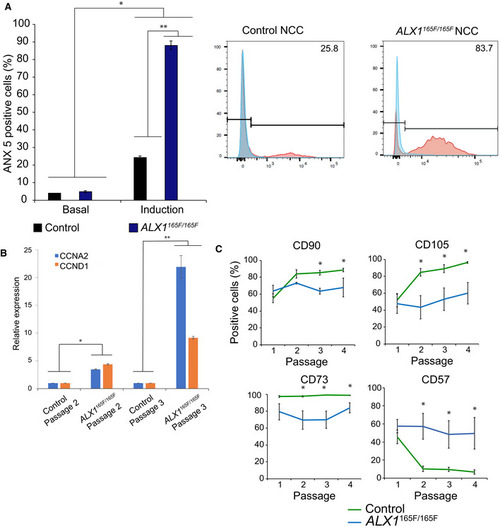
Homozygous Expression levels of cyclins Fluorescence activated cell sorting (FACS) experiments showed that control |
|
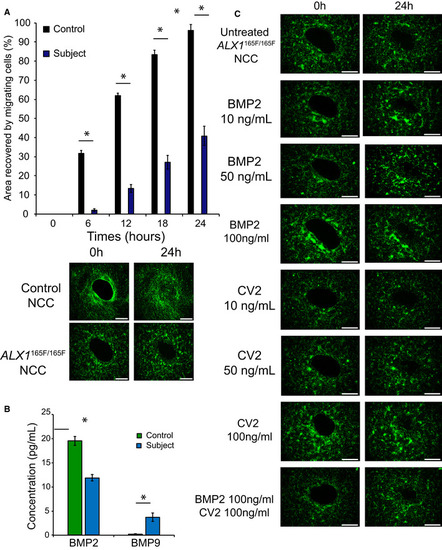
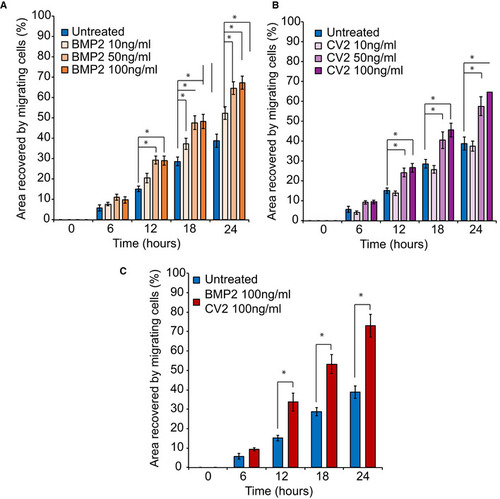
Mutant Multiplex analysis of BMP2 and BMP9 in the supernatant of cultured NCC showed that Addition of soluble BMP2 or CV2, a BMP9 antagonist, to the culture medium could partially rescue the migration defect of |
|
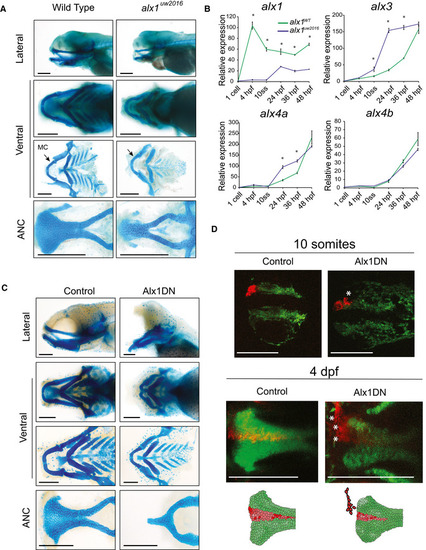
Dissected flatmount wild‐type and Zebrafish Dissected flatmount of zebrafish embryos injected with Alx1DN, after Alcian blue staining. The embryos developed an absence of the frontonasal‐derived median portion of the anterior neurocranium (ANC) and a profound hypoplasia of Meckel's and ventral cartilages. In the most severely affected zebrafish, a nearly abrogated ANC was observed. Scale bar: 200 μm. Lineage tracing experiments in control and Alx1DN mutant embryos revealed aberrant migration of anterior cranial NCC when |
|
Schematic representation of the strategy used to generate iPSC. Blood samples from an unrelated normal individual, unaffected father (subject 1), and two of the affected children (subjects 5 and 6) were processed. Isolated PBMC were infected with Sendai virus, and individual clones were picked 21 days after the infection. Following expansion until passage 10, iPSC were characterized and embryoid bodies were formed by suspension culture for 14 days. The reprogramming process of the PBMC showed that all cells underwent similar morphological changes leading to the formation of iPSC clones by day 21. These clones still displayed embryonic stem cells morphology at passage 10, indicating that the cells are able to self‐renew. All iPSC clones were able to form EBs. One clone of each subject is represented. Scale bar is 400 μm. |
|
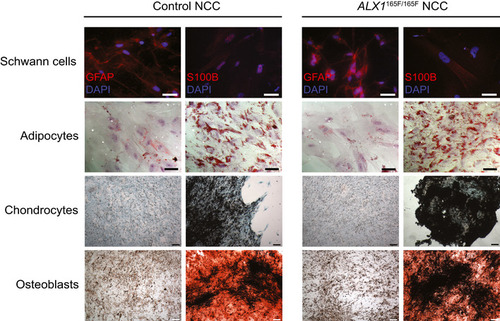
Multilineage differentiation experiments revealed that both control and subject‐derived |
|
10, 50, or 100 ng/ml of soluble BMP2 was added to the culture medium. Surface area analyses and percentages of coverage were measured using ImageJ software (NIH). The data of NCC migration following the treatment with 10, 50, and 100 ng/ml soluble BMP2 are represented as the average of the percentage of closure ± SEM from three independent experiments performed with each clone. To test statistical significance, an ANOVA test was performed. A 10, 50, or 100 ng/ml of soluble CV2 was added to the culture medium. Surface area analyses and percentages of coverage were measured using ImageJ software (NIH). The data of NCC migration following the treatment with 10, 50, and 100 ng/ml soluble CV2 are represented as the average of the percentage of closure ± SEM. Scale bar = 400 μm. *: Significantly different from untreated Recovery of subject‐derived NCC migration in a migration assay following the combined treatment with 100 ng/ml each of soluble BMP2 and CV2. The data are represented as the average of the percentage of closure ± SEM from three independent experiments performed with each clone. To test statistical significance, an ANOVA test was performed. A |
|
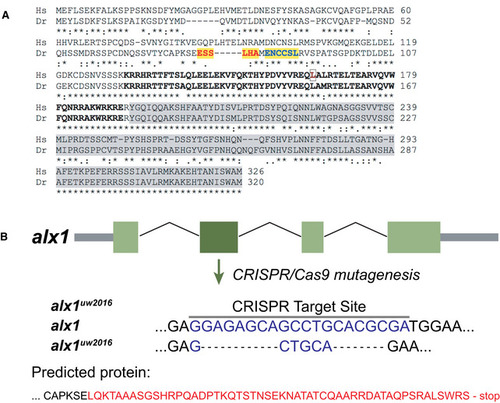
Human ALX1 and zebrafish alx1 protein sequences were obtained from Ensembl and aligned using Clustal Omega ( Schematic diagram shows the effect of the mutant allele resulting from our choice of target site #1. The allele, termed, |
|
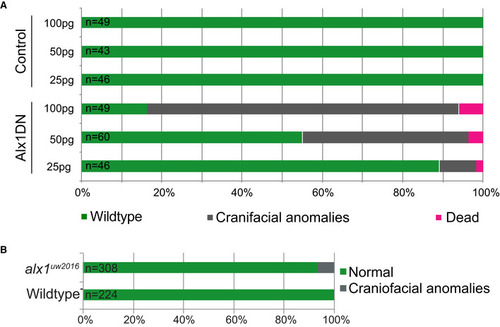
The number of embryos displaying craniofacial phenotypes increased with increasing concentration of Alx1DN mRNA injected into the single cell stage embryo. Overview of the relationship of the results of injections of 25, 50, and 100 pg of control mRNA and Alx1DN mRNA with the outcomes of wild‐type zebrafish (green), a craniofacial phenotype (gray), and dead zebrafish (magenta). The number of embryos displaying craniofacial phenotypes injected with |