- Title
-
Tumour suppressor 15-hydroxyprostaglandin dehydrogenase induces differentiation in colon cancer via GLI1 inhibition
- Authors
- Satapathy, S.R., Topi, G., Osman, J., Hellman, K., Ek, F., Olsson, R., Sime, W., Mehdawi, L.M., Sjölander, A.
- Source
- Full text @ Oncogenesis
|
|
|
|
|
|
|
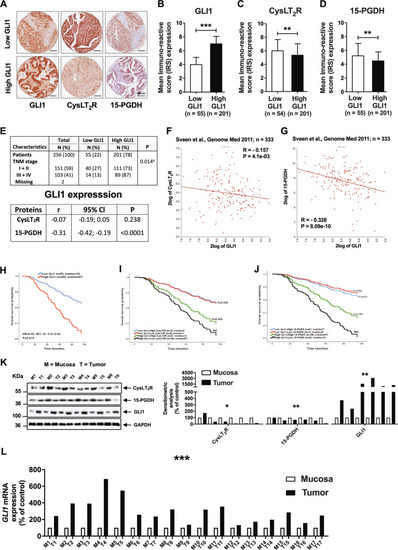
qRT-PCR validation of gene expression in |
|
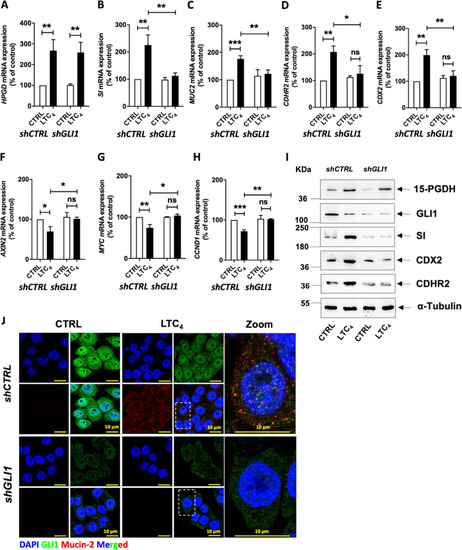
HT-29 cells were either transfected with |
|
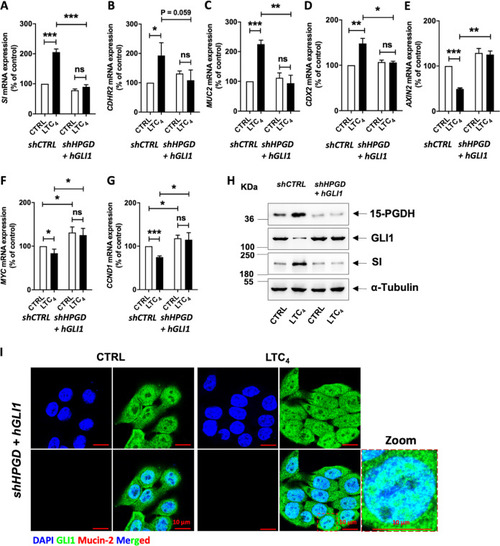
Graphs showing qRT-PCR analysis of |
|
|