- Title
-
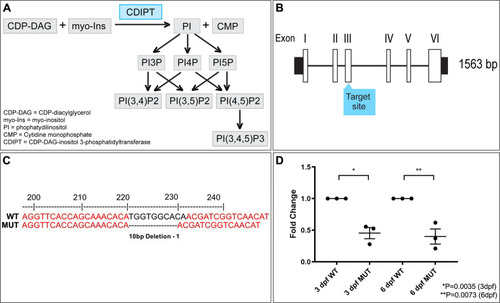
De novo phosphoinositide synthesis in zebrafish is required for triad formation but not essential for myogenesis
- Authors
- Smith, L., Fabian, L., Al-Maawali, A., Noche, R.R., Dowling, J.J.
- Source
- Full text @ PLoS One
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
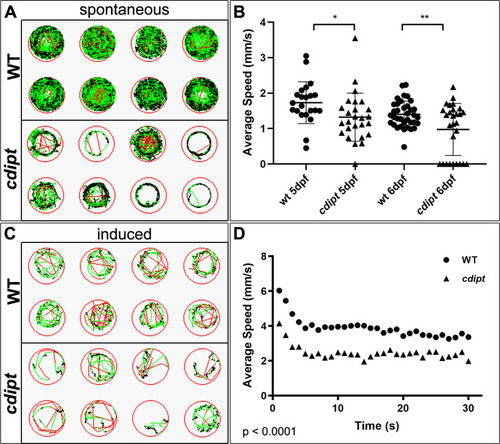
PHENOTYPE:
|
|
PHENOTYPE:
|
|
|
|
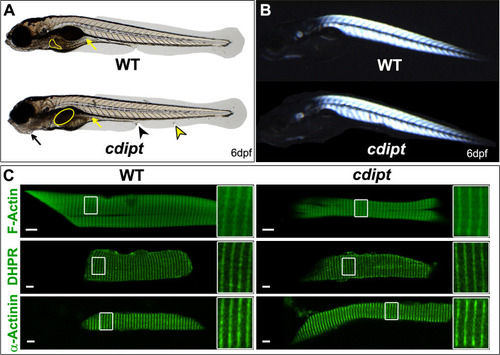
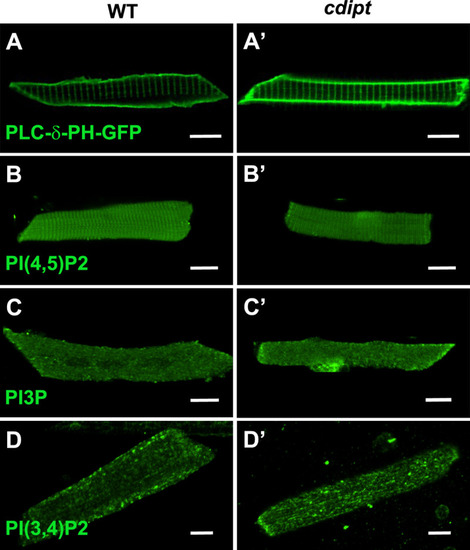
Confocal micrographs showing localization of PIPs is not affected in early larval development of |
|
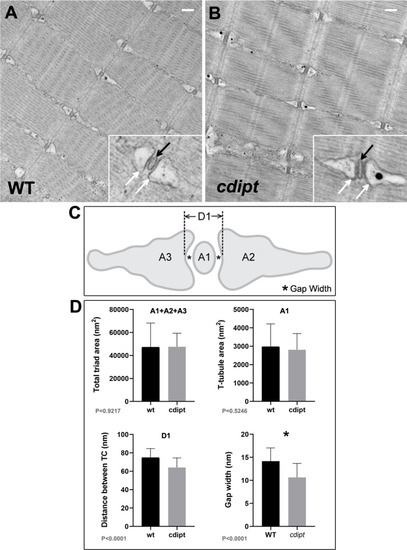
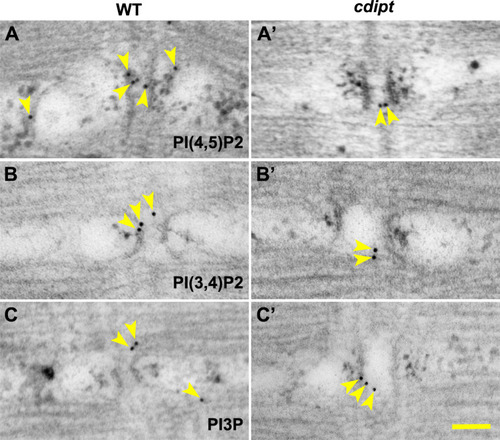
Transmission immunoelectron micrographs showing localization of nanogold-labelled antibodies against |