- Title
-
An optimized QF-binary expression system for use in zebrafish
- Authors
- Burgess, J., Burrows, J.T., Sadhak, R., Chiang, S., Weiss, A., D'Amata, C., Molinaro, A.M., Zhu, S., Long, M., Hu, C., Krause, H.M., Pearson, B.J.
- Source
- Full text @ Dev. Biol.
|
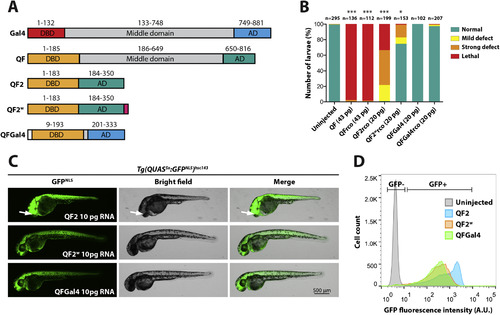
QFGal4 is a non-toxic transcriptional activator in zebrafish. (A) Schematic of Gal4 (S. cerevisiae) and QF (N. crassa) protein domains. QF and Gal4 have a similar domain organization, with an N-terminal DBD, a middle domain, and a C-terminal AD. QF2 lacks the middle domain of QF, and QF2w contains an additional C-terminal polylysine tract to attenuate activity (Riabinina et al., 2015). QFGal4 contains the DNA-binding domain of QF (residues 1–185) fused to the transcriptional activation domain of Gal4 (749–881). QFGal4 also contains an SV40 NLS (PKKKRK) at the N-terminus to facilitate localization to the nucleus. (B) Bar graph showing the relative toxicity of QF and its derivatives in zebrafish embryos. Equimolar amounts of mRNA encoding QF variants were injected into one-cell stage embryos. Full length QF, both original codon usage (n = 136) and recodonized (rco) versions (n = 112), was strongly toxic with most embryos exhibiting lethality during the blastula stage (98% and 98%, respectively). QF2 recodonized (n = 199) was also toxic, with the majority of embryos exhibiting either lethality (35%) or strong developmental defects (44%). QF2w recodonized (n = 153) was well tolerated with 71% of embryos appearing healthy. QFGal4, both original codon usage (n = 102) and QF recodonized (n = 207), was well tolerated (100% and 98% normal, respectively). Injected embryos were scored as follows: mild defects included heart edema and/or mild tail kinking; severe defects included gross developmental defects, such as failure to form embryonic structures like the eye; and lethality was scored as embryos that did not proceed past the blastula stage. QF, QFrco, QF2rco and QF2wrco were significantly different from uninjected control embryos, while no significant difference was detected for either QFGal4 or QFGal4rco as compared to uninjected controls. (C) Epifluorescence micrographs showing the in vivo activity of either QF2, QF2w or QFGal4 as determined by RNA injection into a fourth-generation Tg(QUAS5x:GFPNLS)hsc143 reporter line. 10 pg of mRNA encoding either QF2 (top panel), QF2w (middle panel) or QFGal4 (lower panel) was injected at the one-cell stage and larvae were imaged at 2 dpf. Tg(QUAS5x:GFPNLS)hsc143 larvae injected with QF2 mRNA exhibited strong, ubiquitous GFPNLS expression, although larvae exhibited developmental defects, including misplaced eyes (arrow). Tg(QUAS5x:GFPNLS)hsc143 larvae injected with either QF2w or QFGal4 mRNA also exhibited ubiquitous GFPNLS expression, although signal appeared weaker than embryos injected with QF2 mRNA. (D) Plot showing relative GFPNLS fluorescence intensity of 2 dpf disassociated Tg(QUAS5x:GFPNLS)hsc143 transgenic embryos injected with 10 pg mRNA encoding either QF2, QF2w or QFGal4 mRNA. QF2 (blue) resulted in the strongest activation of a Tg(QUAS5x:GFPNLS)hsc143 reporter, followed by QF2w (orange) and QFGal4 (green). 30,000 events per sample are shown. |
|
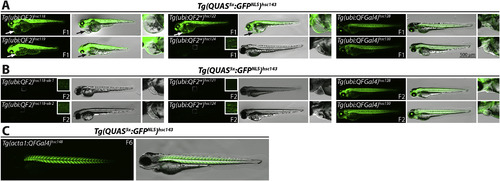
Generation of a stably-integrated, ubiquitously-expressed QFGal4 driver line. (A) Epifluorescence micrographs showing GFPNLS expression from a Tg(QUAS5x:GFPNLS)hsc143 reporter line crossed to either Tg(ubi:QF2) (left column), Tg(ubi:QF2w) (middle column), or Tg(ubi:QFGal4) (right column) transgenic F0 founder fish. A magnified view of the heart chamber is shown to the right of each merged panel. Tg(ubi:QF2) and Tg(ubi:QF2w) transgenic lines exhibited the strongest activation of the GFPNLS reporter, but also had developmental defects, including heart edema (arrow and dashed line outlining edema in insets). Notably, defects caused by QF2 and QF2w expression were also observed in the absence of the Tg(QUAS5x:GFPNLS)hsc143 transgene (not shown). Most Tg(ubi:QF2w) F1 larvae exhibited strong GFPNLS expression, except progeny from Tg(ubi:QF2w)hsc124, which showed weak variegated GFPNLS expression when crossed a Tg(QUAS5x:GFPNLS)hsc143 reporter line. Tg(ubi:QFGal4) lines showed robust and ubiquitous activation of a GFPNLS reporter line, with no observable toxic effects during larval development. Note that each F1 larvae shown is derived from an independent founder. All larvae shown were imaged at 3 dpf. (B) Epifluorescence micrographs showing GFPNLS expression from a Tg(QUAS5x:GFPNLS)hsc143 reporter line crossed to first generation Tg(ubi:QF2) (left column), Tg(ubi:QF2w) (middle column) or Tg(ubi:QFGal4) transgenic fish (right column). A magnified view of the heart chamber is shown to the right of each merged panel. Tg(ubi:QF2)hsc118 and Tg(ubi:QF2w)hsc124 F2 larvae exhibited weak, mosaic GFPNLS expression (see insets), and Tg(ubi:QF2w)hsc121 exhibited no detectable GFPNLS expression when crossed to a Tg(QUAS5x:GFPNLS)hsc143 reporter line. In contrast, Tg(ubi:QFGal4) F2 larvae exhibited strong, ubiquitous GFPNLS expression. No heart edema was evident in F2 larvae for any of the transgenic lines (see inset panels). Each larvae shown is an F2 established from an independent founder, and all larvae were imaged at 3 dpf. (C) Epifluorescence micrographs showing GFPNLS expression from a Tg(QUAS5x:GFPNLS)hsc143 reporter line crossed to a muscle-specific driver line Tg(acta1:QFGal4)hsc148. Image shows a 3 dpf larvae. |
|
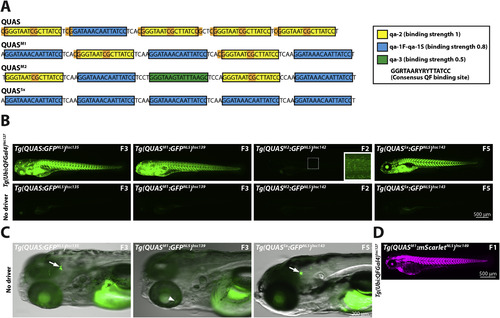
Optimization of QUAS binding sites. (A) Schematic depicting QUAS binding sites for various QUAS elements. The original QUAS element contains 4 copies of qa-2 (yellow box) and one copy of qa-1F-qa-1S (blue box) (Potter et al., 2010). CpG dinucleotides are highlighted in orange. Note that CpG dinuclotides are present in linker regions and the non-consensus nucleotides of the qa-2 binding site. To reduce CpG content, and increase variability, we generated QUAS mixed-1 (QUASM1), QUAS mixed-2 (QUASM2) and QUAS5x. These new QUAS elements contain fewer copies of the qa-2 binding site to reduce CpG content. In addition, QUASM2 contains the qa-3 binding site (green box), not present in the original QUAS element. QUASM1, QUASM2, and QUAS5x contain, four, two and no CpG dinucleotides, respectively. Neurospora crassa QF-binding sites, as well as relative affinity of the QF DBD to each binding site, was described previously (Baum et al., 1987). Yellow box: qa-2 (relative QF DBD binding strength of 1); Blue box: qa-1F-qa-1S (relative binding strength of 0.8); Green box: qa-3 (relative binding strength of 0.5). The consensus QF binding site is shown in the boxed inset. (B) Epifluorescence micrographs showing representative GFPNLS expression from either Tg(QUAS:GFPNLS)hsc135, Tg(QUASM1:GFPNLS)hsc139, Tg(QUASM2:GFPNLS)hsc142 or Tg(QUAS5x:GFPNLS)hsc143 reporter lines crossed to a fourth generation Tg(ubi:QFGal4)hsc127 driver line. QUAS5, QUASM1 and QUAS5x elements exhibited comparable in vivo activity, as assessed by GFPNLS reporter expression. In contrast, the QUASM2 element showed weak in vivo activity. All larvae shown are 5 dpf larvae, and the generation of the reporter line is indicated. (C) Epifluorescence micrographs showing leaky GFPNLS expression from QUAS and QUAS5x reporter lines in the absence of a QF driver line. Leaky GFPNLS expression was observed in the brain for Tg(QUAS:GFPNLS)hsc135 and Tg(QUAS5x:GFPNLS)hsc143, but not for Tg(QUASM1:GFPNLS)hsc139 (arrow). Conversely, faint variegated GFPNLS expression was observable in the retina of Tg(QUASM1:GFPNLS)hsc139 (arrowhead). Leaky expression for QUAS, QUAS5x and QUASM1 was evident in multiple, independently derived transgenic lines (see Supplemental Fig. 2B). All larvae were treated with PTU to block pigmentation and were imaged at 5 dpf. (D) Epifluorescence micrograph showing ubiquitous mScarletNLS expression from a Tg(QUASM1:mScarletNLS)hsc149 reporter line crossed to a Tg(ubi:QFGal4)hsc127 driver line. An F1 larvae imaged at 3 dpf is shown. |
|
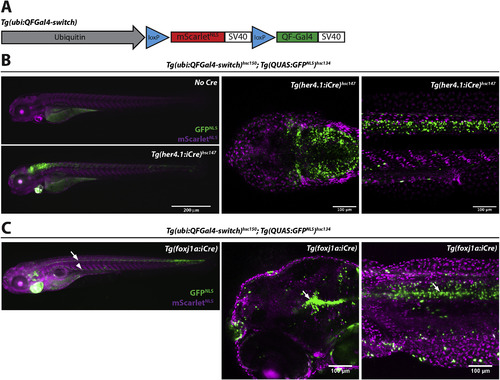
Lineage tracing using ubi:QFGal4-switch. (A) Schematic depicting the organization of a Cre-inducible ubi:QFGal4-switch construct. In the absence of Cre, the ubiquitin B promoter drives expression of mScarletNLS. In the presence of Cre, the loxP-mScarletNLS-loxP expression cassette is excised, resulting in QFGal4 expression. Tg(ubi:QFGal4-switch) transgenic fish can be combined with any existing QUAS reporter and Cre line to create specifically and permanently marked cells for lineage tracing experiments. (B) Fluorescence micrographs showing GFPNLS expression in her4:iCre expressing cells. Epifluorescence micrographs of Tg(ubi:QFGal4-switch)hsc150; Tg(QUAS:GFPNLS)hsc134 double transgenic larvae without Cre (top left panel), or crossed to a Tg(her4.1:iCre)hsc147 transgenic line (bottom left panel). Larvae positive for all three transgenes exhibit robust GFPNLS expression in glial cells of the brain and spinal cord. Right panels: Confocal micrographs showing GFPNLS expression in the brain (coronal section - middle panel) and spinal cord (lateral view - right panel). Note that non-switched cells are marked by mScarletNLS. 4 dpf larvae are shown. (C) Fluorescence micrographs showing GFPNLS expression in foxj1a:iCre expressing cells. Epifluorescence micrographs of Tg(ubi:QFGal4-switch)hsc150; Tg(QUAS:GFPNLS)hsc134 double transgenic larvae crossed to a Tg(foxj1a:iCre) transgenic line and imaged at 5 dpf (left panel). GFPNLS expression is visible along the spinal cord (arrow) and pronephros (arrowhead). Confocal micrographs showing GFPNLS expression in the brain ventricles (lateral view - middle panel) and spinal cord (lateral view - right panel) (Van Gennip et al., 2018).
Construct:
Tg(QUASM1-E1B:mScarlet-NLS,myl7:mRFP)
|
|
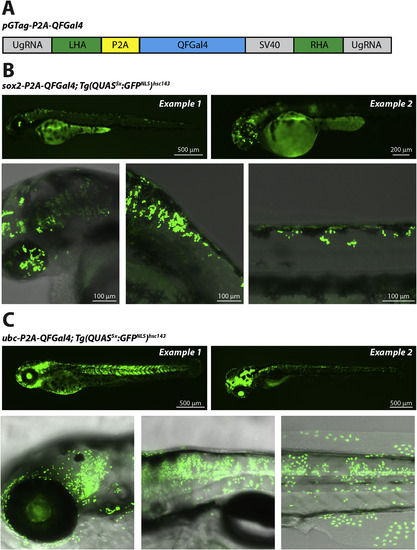
Tagging endogenous loci with QFGal4. (A) Schematic showing the pGTag:QFGal4 gene tag construct. The P2A:QFGal4:SV40 expression cassette is flanked by left and right homology arms (LHA and RHA, respectively). In addition, the vector contains universal gRNA (UgRNA) target sequences, that can be cut in vivo by coinjection with Cas9 and UgRNA (modified from (Wierson et al., 2020)). (B) Confocal fluorescence micrographs showing GFPNLS expression in sox2-QFGal4 knock-in tagged cells. Tg(QUAS5x:GFPNLS)hsc143 transgenic lines were injected with pGTag:QFGal4, sox2 TALEN mRNA, as well as UgRNA and Cas9 protein to cut the pGTag vector in vivo. Successful QFGal4 integration at the sox2 locus was visualized by Tg(QUAS5x:GFPNLS)hsc143 reporter activity. Mosaic GFPNLS expression was visible throughout the spinal cord and retina in 3 dpf larvae (top two panels). GFPNLS labeling was visible in the retina and brain (lower left panel), hindbrain (middle lower panel) and spinal cord (lower right panel). (C) Fluorescence micrographs showing GFPNLS expression in ubiquitin C (ubc) expressing cells tagged with QFGal4. Tg(QUAS5x:GFPNLS)hsc143 transgenic lines were injected with pGTag:QFGal4, ubc gRNA, UgRNA and Cas9 protein. Epifluorescence micrographs show mosaic GFPNLS expression visible throughout the larval body, consistent with broad integration of QFGal4 at the ubc locus (3 dpf, top left panel and 5 dpf, top right panel). Confocal micrographs showing GFPNLS expression in a range of cells, including brain and retina (lower left panel), trunk (lower middle panel) and spinal cord, muscle, epidermal cells (lower right panel).
Construct:
Tg(QUAS5X-E1B:GFP-NLS,myl7:mRFP)
|
|
Supplemental figure 1. |
|
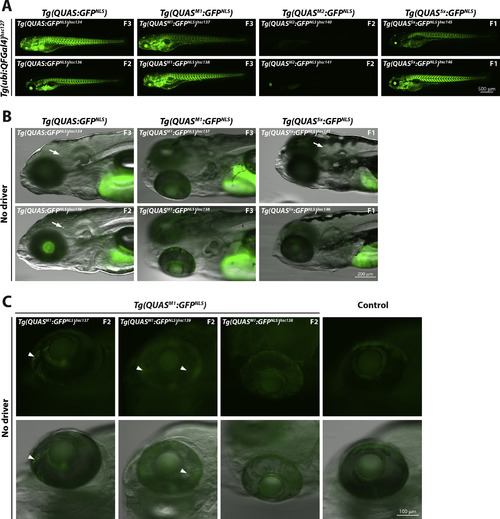
Supplemental figure 2. |
|
Supplemental figure 3. |
Reprinted from Developmental Biology, 465(2), Burgess, J., Burrows, J.T., Sadhak, R., Chiang, S., Weiss, A., D'Amata, C., Molinaro, A.M., Zhu, S., Long, M., Hu, C., Krause, H.M., Pearson, B.J., An optimized QF-binary expression system for use in zebrafish, 144-156, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.