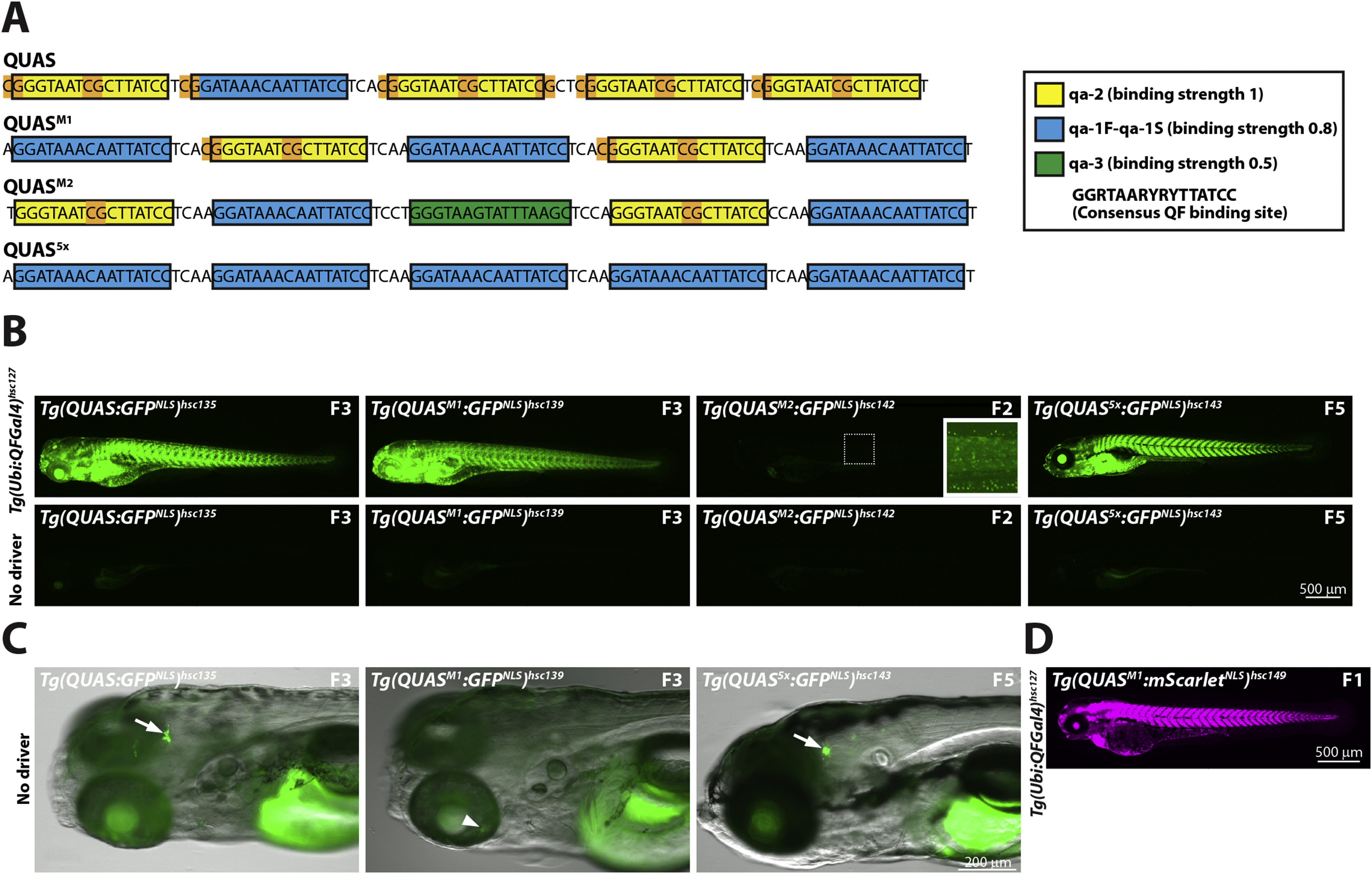
Fig. 3 Optimization of QUAS binding sites. (A) Schematic depicting QUAS binding sites for various QUAS elements. The original QUAS element contains 4 copies of qa-2 (yellow box) and one copy of qa-1F-qa-1S (blue box) (Potter et al., 2010). CpG dinucleotides are highlighted in orange. Note that CpG dinuclotides are present in linker regions and the non-consensus nucleotides of the qa-2 binding site. To reduce CpG content, and increase variability, we generated QUAS mixed-1 (QUASM1), QUAS mixed-2 (QUASM2) and QUAS5x. These new QUAS elements contain fewer copies of the qa-2 binding site to reduce CpG content. In addition, QUASM2 contains the qa-3 binding site (green box), not present in the original QUAS element. QUASM1, QUASM2, and QUAS5x contain, four, two and no CpG dinucleotides, respectively. Neurospora crassa QF-binding sites, as well as relative affinity of the QF DBD to each binding site, was described previously (Baum et al., 1987). Yellow box: qa-2 (relative QF DBD binding strength of 1); Blue box: qa-1F-qa-1S (relative binding strength of 0.8); Green box: qa-3 (relative binding strength of 0.5). The consensus QF binding site is shown in the boxed inset. (B) Epifluorescence micrographs showing representative GFPNLS expression from either Tg(QUAS:GFPNLS)hsc135, Tg(QUASM1:GFPNLS)hsc139, Tg(QUASM2:GFPNLS)hsc142 or Tg(QUAS5x:GFPNLS)hsc143 reporter lines crossed to a fourth generation Tg(ubi:QFGal4)hsc127 driver line. QUAS5, QUASM1 and QUAS5x elements exhibited comparable in vivo activity, as assessed by GFPNLS reporter expression. In contrast, the QUASM2 element showed weak in vivo activity. All larvae shown are 5 dpf larvae, and the generation of the reporter line is indicated. (C) Epifluorescence micrographs showing leaky GFPNLS expression from QUAS and QUAS5x reporter lines in the absence of a QF driver line. Leaky GFPNLS expression was observed in the brain for Tg(QUAS:GFPNLS)hsc135 and Tg(QUAS5x:GFPNLS)hsc143, but not for Tg(QUASM1:GFPNLS)hsc139 (arrow). Conversely, faint variegated GFPNLS expression was observable in the retina of Tg(QUASM1:GFPNLS)hsc139 (arrowhead). Leaky expression for QUAS, QUAS5x and QUASM1 was evident in multiple, independently derived transgenic lines (see Supplemental Fig. 2B). All larvae were treated with PTU to block pigmentation and were imaged at 5 dpf. (D) Epifluorescence micrograph showing ubiquitous mScarletNLS expression from a Tg(QUASM1:mScarletNLS)hsc149 reporter line crossed to a Tg(ubi:QFGal4)hsc127 driver line. An F1 larvae imaged at 3 dpf is shown.
Reprinted from Developmental Biology, 465(2), Burgess, J., Burrows, J.T., Sadhak, R., Chiang, S., Weiss, A., D'Amata, C., Molinaro, A.M., Zhu, S., Long, M., Hu, C., Krause, H.M., Pearson, B.J., An optimized QF-binary expression system for use in zebrafish, 144-156, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.