- Title
-
Translating GWAS-identified loci for cardiac rhythm and rate using an in vivo image- and CRISPR/Cas9-based approach
- Authors
- von der Heyde, B., Emmanouilidou, A., Mazzaferro, E., Vicenzi, S., Höijer, I., Klingström, T., Jumaa, S., Dethlefsen, O., Snieder, H., de Geus, E., Ameur, A., Ingelsson, E., Allalou, A., Brooke, H.L., den Hoed, M.
- Source
- Full text @ Sci. Rep.
|
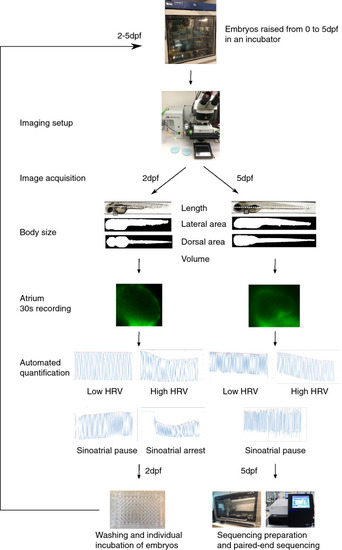
Overview of the experimental pipeline showing the image-based acquisition and post-processing of images and samples. |
|
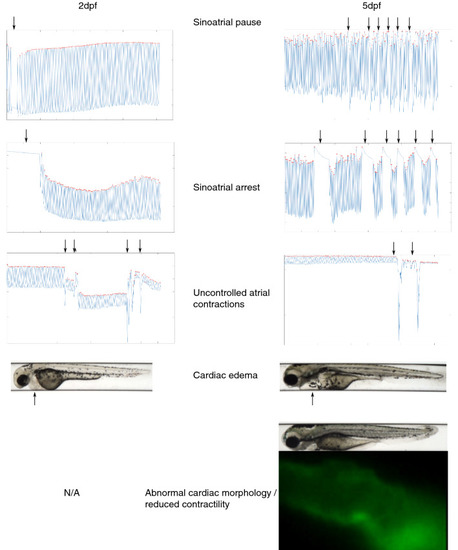
Visualization of cardiac rhythmic or other abnormalities, highlighted by arrows. Sinoatrial pauses were defined as the atrium ceasing to contract for > 3 × the median inter beat interval of the embryo. A sinoatrial arrest was defined as an event where the atrium stopped contracting for > 2 s. Abnormal cardiac morphology was defined as the atrium appearing as a tube-like structure, and impaired cardiac contractility was defined as the atrium vibrating rather than contracting. |
|
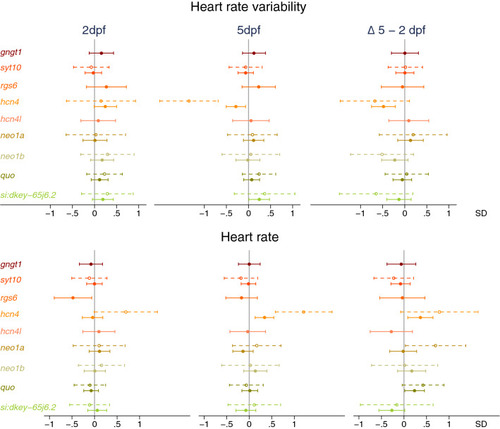
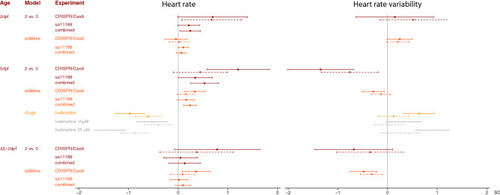
Effect of CRISPR/Cas9-induced mutations in candidate genes on (change in) heart rate variability (top) and heart rate (bottom) at 2 days post-fertilization (dpf, n = 234) and 5dpf (n = 285). Full dots and solid whiskers show the effect size and 95% confidence interval (CI) for each additional mutated allele, weighted by the mutation’s predicted effect on protein function. Open dots and dotted whiskers indicate the effect and 95% CI for nonsense mutations in both alleles vs. no CRISPR/Cas9-induced mutations. Effects were adjusted for the weighted number of mutated alleles in the other targeted genes, as well as for time of day (fixed factors), with embryos nested in batches (random factor). |
|
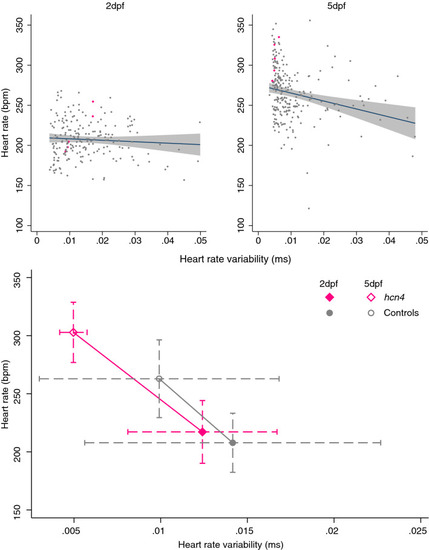
Association of heart rate variability and heart rate at 2- and 5-days post-fertilization (dpf) and effect of nonsense mutations in both |
|
The effect of mutations in |
|
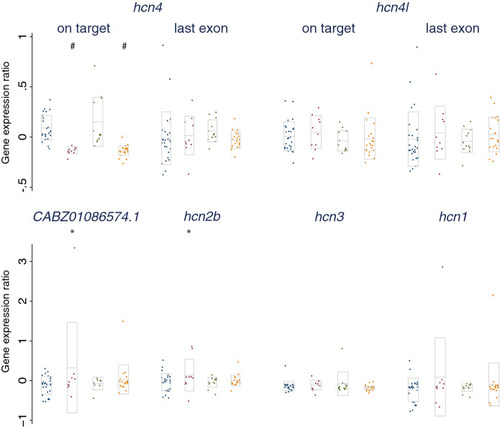
qRT-PCR results for the expression of transcripts with high (> 75%) sequence similarity to the main zebrafish |