|
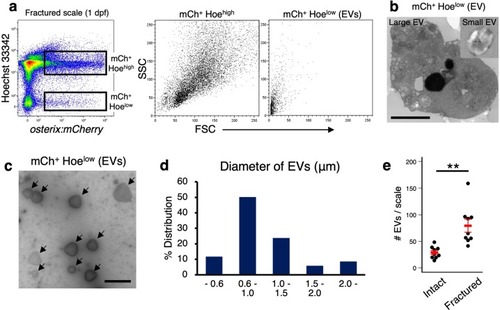
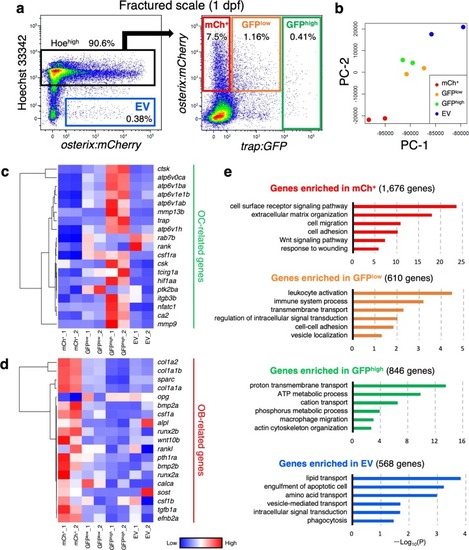
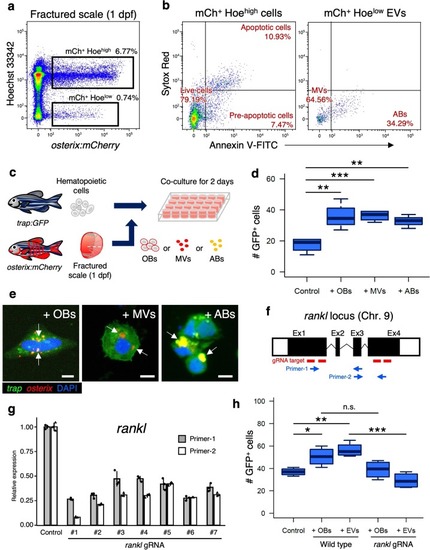
OB-derived EVs promote OC differentiation via Rankl signaling.a Representative flow cytometric analysis of cells in scales at 1 day post-fracture (dpf) from a osterix:mCherry single-transgenic animal. Gated regions indicate the mCh+ Hoehigh cell fraction and mCh+ Hoelow EV fraction. b mCh+ Hoehigh cells and mCh+ Hoelow EVs were displayed in an Annexin-V-FITC vs. Sytox Red dot plot. mCh+ Hoehigh cells were subdivided into three fractions, “live”, “pre-apoptotic”, and “apoptotic”, whereas mCh+ Hoelow EVs were divided into two fractions, “microvesicle” (MV) and “apoptotic body” (AB). c Schematic diagram of in vitro cell culture assays. Kidney marrow cells (KMCs) from trap:GFP single-transgenic zebrafish were co-cultured with OBs, MVs, or ABs from fractured scales of osterix:mCherry single-transgenic zebrafish in a fibronectin-coated plate. At 2 days of co-culture, the number of trap:GFP+ cells was counted in each well. Non-co-cultured KMCs were used as a control. d The average number of trap:GFP+ cells in each type of wells. Error bars, s.d. (n = 4 for each group). e Representative images of trap:GFP+ cells co-cultured with OBs (left panel), MVs (middle panel), or ABs (right panel). trap:GFP+ cells contained OB-derived EVs in the cytoplasm (arrows). Bars, 5 μm. f Schematic diagram of zebrafish rankl locus. gRNA target sites and primer recognition sites are shown in red bars and blue arrows, respectively. g qPCR analysis of rankl in the adult fin of wild type control and rankl gRNA-injected animals. Results from seven individual gRNA-injected animals are shown. Data are mean ± s.d. from three independent experiments. h The average number of trap:GFP+ cells in non-co-cultured KMCs (control), or KMCs co-cultured with OBs or EVs from wild type or rankl gRNA-injected animals. Error bars, s.d. (n = 4 for each group); n.s., no significance; *p < 0.05; **p < 0.01; ***p < 0.001 by one-way ANOVA followed by Dunnett’s test. Experiments were performed twice with four biological replicates in each group d, h.
|