- Title
-
Patterns of spon1b:GFP expression during early zebrafish brain development
- Authors
- Agudelo-Dueñas, N., Forero-Shelton, M., Zhdanova, I.V., Akle, V.
- Source
- Full text @ BMC Res. Notes
|
|
|
|
|
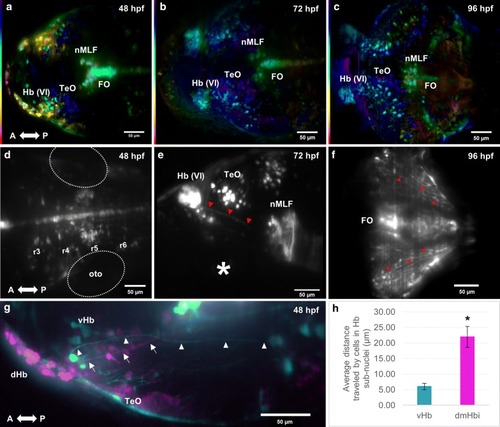
Development of the habenula from 48 to 73 hpf. Development of the Hb complex followed through time-lapse imaging. Cells within the dorsal habenula (dHb) aggregate progressively during development from an elongated shape to form a nucleus as indicated by the white arrowheads. The dHb is observed to be located more dorsally with respect to the vHb. Images from |