- Title
-
Notch-Mediated Determination of Hair-Bundle Polarity in Mechanosensory Hair Cells of the Zebrafish Lateral Line
- Authors
- Jacobo, A., Dasgupta, A., Erzberger, A., Siletti, K., Hudspeth, A.J.
- Source
- Full text @ Curr. Biol.
|
Organization of Neuromasts in the Zebrafish’s Lateral Line (A) A schematic drawing of a 4 dpf larva depicts neuromasts (green) of the posterior lateral line. Additional, unshown neuromasts adorn the anterior portion of the larva. (B) A diagrammatic surface view of a single neuromast shows hair cells (green) separated by supporting cells (black) and surrounded by mantle cells (gray). (C) A schematic section depicts the sensitivity of oppositely oriented hair cells to water flow. (D) In a micrograph of the upper surface of a neuromast, spectrin (green) marks the cuticular plates of hair cells. Immunolabeling reveals that the core PCP protein Vangl2 (orange) occurs consistently at the posterior cellular boundaries. (E) A polar plot of average Vangl2 labeling from 502 hair cells quantitates the effect in (D). This and each of the subsequent polar plots represents the average intensity of labeling, normalized to the maximum value, as a function of angular position with respect to the cells’ centers. (F) In a developing neuromast, Emx2 (magenta) is expressed in the nuclei of three mature hair cells (green dashed lines on the left). In the distribution of actin-GFP (green) at the cellular apices, the dark spots locate the kinocilia and indicate that those cells are sensitive to caudad stimulation. Three other mature hair cells (green dashed lines on the right) have an opposite orientation. The apices of two immature hair cells (gray dashed lines) are indicated by arrowheads. (G) The polarities of hair cells from 20 neuromasts (top) in 4 dpf larvae are equally divided between caudad (Cd; 135 cells) and rostrad (Rd; 141 cells). The former cells express significantly higher levels of Emx2 (bottom). Scale bars, 3 μm. Means ± SEM; ∗∗∗p < 0.001. See also Figure S1. EXPRESSION / LABELING:
|
|
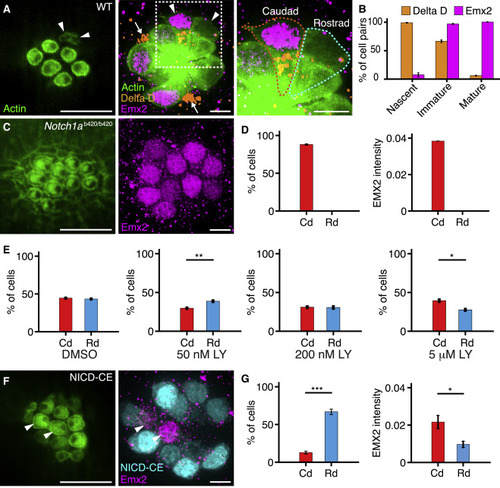
Experimental Perturbations of the Notch Signaling Pathway (A) An apical view of a 4 dpf wild-type neuromast (left) reveals three mature pairs of hair cells and one immature pair (arrowheads). A view at the nuclear level (center) shows immunolabeling of the same neuromast for Emx2 (magenta) and Delta D (orange). An enlargement of the boxed area (right) includes outlines of the two immature hair cells. Hair-cell progenitors also express Delta D (arrows). (B) Delta D is expressed in at least one hair cell of each pair at earlier stages of development than Emx2 (51 nascent pairs, 101 immature pairs, 259 mature pairs). (C) In a Notch1ab420/b420 mutant, hair cells show a caudad polarization (left) and uniformly express Emx2 (right). (D) These effects are consistent in 536 hair cells from 57 neuromasts. (E) The γ-secretase inhibitor LY411575 creates a polarity bias that lies in the rostrad direction for low concentrations but becomes caudad at higher concentrations. The measurements were made as follows: for 50 nM compound, 160 caudad and 205 rostrad hair cells in 37 neuromasts; for 200 nM compound, 47 caudad and 48 rostrad hair cells in 12 neuromasts; and for 5 μM compound, 164 caudad and 114 rostrad hair cells in 19 neuromasts. (F) In an NICD-CE transgenic animal, most hair cells are polarized in the rostrad direction (left). Occasional hair cells (arrowheads) fail to express NICD-CE; they instead contain Emx2 (magenta, right) and adopt a caudad sensitivity. (G) The effect is quantified for 68 cells from 21 neuromasts. Scale bars, 5 μm. Means ± SEM; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05. See also Figure S3 and Videos S1, S2, S3, and S4. |
|
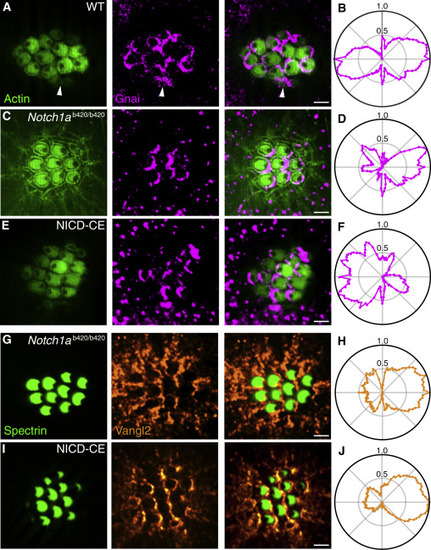
Parallel Action of Notch and PCP (A) In an apical view of a wild-type neuromast, hair-cell polarity is delineated by actin-GFP (green, left) and Gnai is immunolabeled (magenta, center). As shown in the merged view (right), Gnai already displays a polarized location in immature hair cells (arrowheads). (B) In the average radial intensity profile of Gnai from 239 hair cells of both polarities, caudad and rostrad cells create symmetrical lobes on the two halves of the apical surface. (C) Notch1ab420/b420 hair cells display a consistently caudad orientation with crescents of Gnai at their posterior boundaries. (D) A polar plot from 81 hair cells confirms the asymmetrical distribution of Gnai. (E) NICD-CE hair cells are consistently polarized in the rostrad direction and bear Gnai at their anterior edges. (F) A polar plot from 35 hair cells confirms the asymmetry of Gnai distribution. (G) Immunolabeling for spectrin (green, left) shows that Notch1ab420/b420 hair cells are caudad polarized; labeling for Vangl2 (orange, center) displays its distribution at the posterior edges of the cells. (H) A polar plot from 155 hair cells confirms the asymmetrical distribution of Vangl2. (I) Although NICD-CE hair cells are consistently rostrad-polarized, they also bear Vangl2 at their posterior edges. (J) A polar plot from 235 hair cells confirms the asymmetry. Scale bars, 2 μm. See also Figure S4. PHENOTYPE:
|
|
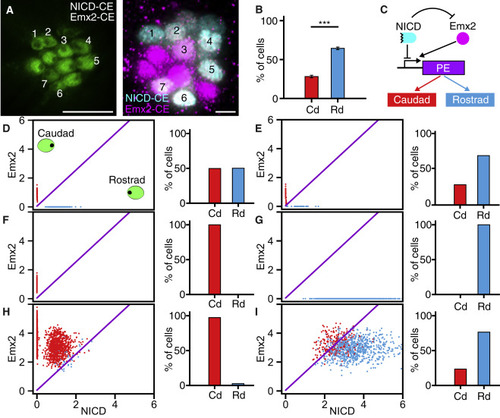
Modeling of the Regulatory Network Underlying Hair-Cell Polarity (A) In an apical view, numerals mark the hair cells in a larva constitutively expressing both NICD (cyan) and Emx2 (magenta). (B) In larvae expressing both proteins, 334 hair cells from 50 neuromasts display a strong bias toward rostrad polarity. (C) In the proposed regulatory network, Notch and Emx2 control hair-cell polarity through a downstream polarity effector (PE), which might be a single protein or a more complex system. (D–I) The panels display the steady-state expression of Emx2 and NICD for multiple simulations of hair-cell pairs of (D) wild-type, (E) DAPT-treated, (F) Notch1a−/−, (G) NICD-CE, (H) Emx2-CE, and (I) NICD-CE/Emx2-CE neuromasts. In each panel, the oblique purple line demarcates the threshold for switching of the polarity effector. Scale bars, 5 μm. Means ± SEM; ∗∗∗p < 0.001. See also Figure S5 and Video S5. |