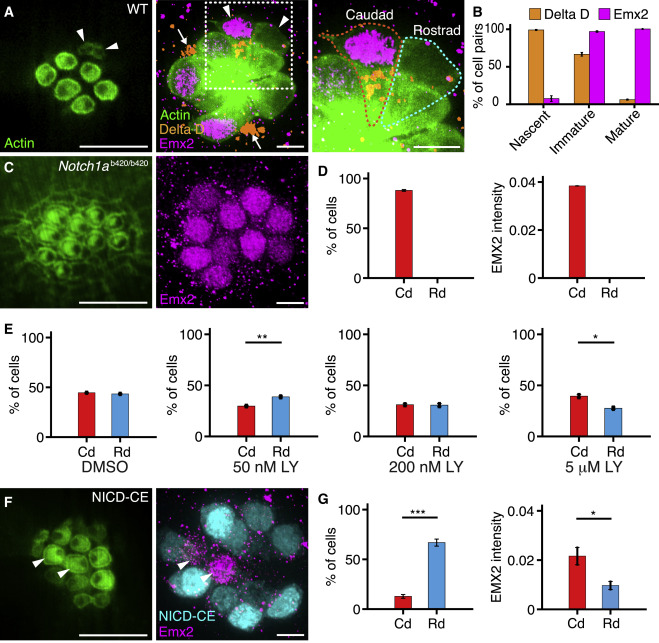
Fig. 3 Experimental Perturbations of the Notch Signaling Pathway (A) An apical view of a 4 dpf wild-type neuromast (left) reveals three mature pairs of hair cells and one immature pair (arrowheads). A view at the nuclear level (center) shows immunolabeling of the same neuromast for Emx2 (magenta) and Delta D (orange). An enlargement of the boxed area (right) includes outlines of the two immature hair cells. Hair-cell progenitors also express Delta D (arrows). (B) Delta D is expressed in at least one hair cell of each pair at earlier stages of development than Emx2 (51 nascent pairs, 101 immature pairs, 259 mature pairs). (C) In a Notch1ab420/b420 mutant, hair cells show a caudad polarization (left) and uniformly express Emx2 (right). (D) These effects are consistent in 536 hair cells from 57 neuromasts. (E) The γ-secretase inhibitor LY411575 creates a polarity bias that lies in the rostrad direction for low concentrations but becomes caudad at higher concentrations. The measurements were made as follows: for 50 nM compound, 160 caudad and 205 rostrad hair cells in 37 neuromasts; for 200 nM compound, 47 caudad and 48 rostrad hair cells in 12 neuromasts; and for 5 μM compound, 164 caudad and 114 rostrad hair cells in 19 neuromasts. (F) In an NICD-CE transgenic animal, most hair cells are polarized in the rostrad direction (left). Occasional hair cells (arrowheads) fail to express NICD-CE; they instead contain Emx2 (magenta, right) and adopt a caudad sensitivity. (G) The effect is quantified for 68 cells from 21 neuromasts. Scale bars, 5 μm. Means ± SEM; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05. See also Figure S3 and Videos S1, S2, S3, and S4.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Curr. Biol.