- Title
-
Cancer-associated fibroblast-derived Gremlin 1 promotes breast cancer progression
- Authors
- Ren, J., Smid, M., Iaria, J., Salvatori, D.C.F., van Dam, H., Zhu, H.J., Martens, J.W.M., Ten Dijke, P.
- Source
- Full text @ Breast Cancer Res.
|
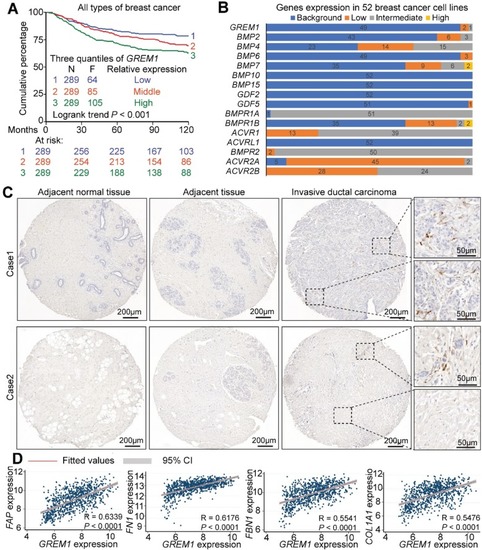
Stromal expression of |
|
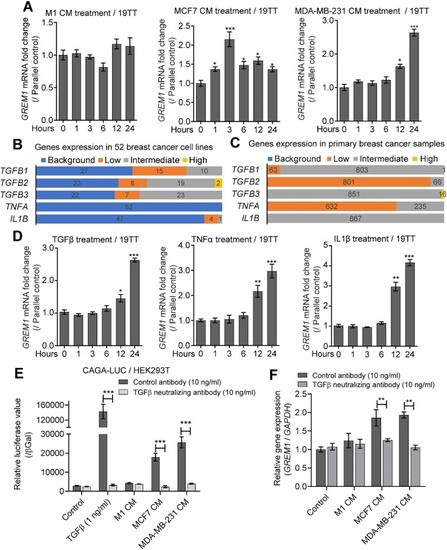
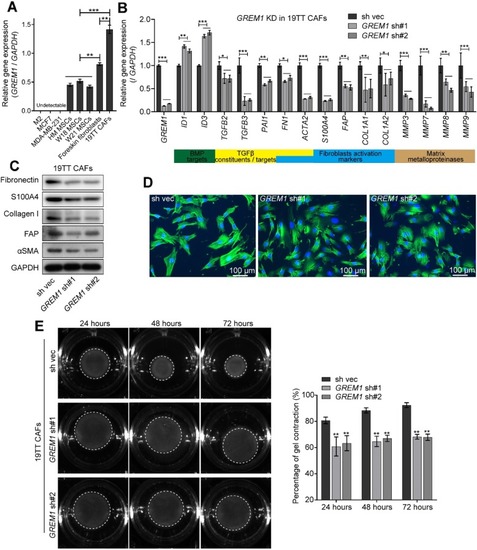
TGFβ secreted by breast cancer cells and inflammatory cytokines induce |
|
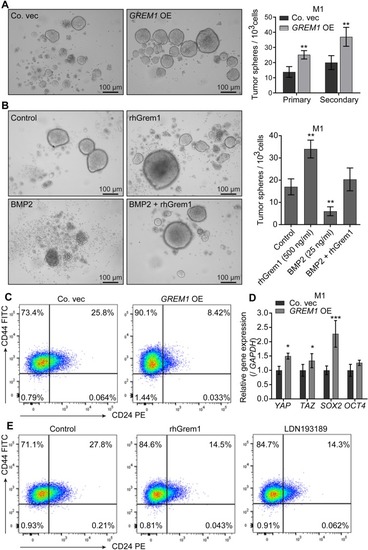
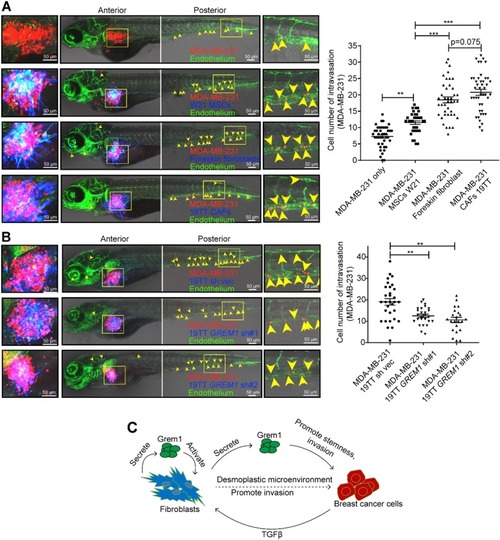
Grem1 maintains stemness in M1 cells. |
|
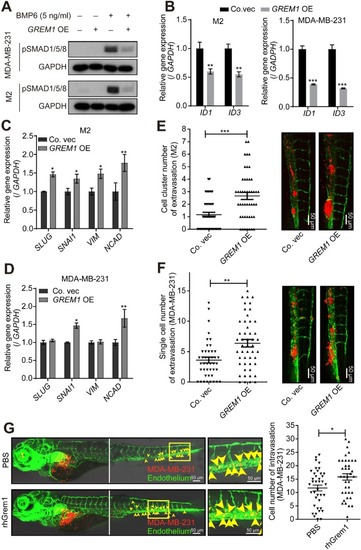
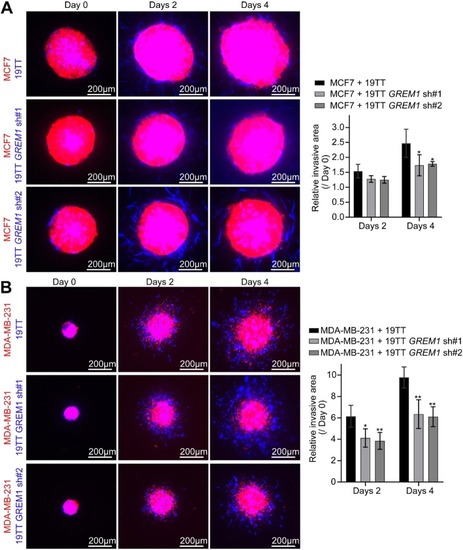
Ectopic expression of |
|
|
|
|
|
|