- Title
-
A critical role of VMP1 in lipoprotein secretion
- Authors
- Morishita, H., Zhao, Y.G., Tamura, N., Nishimura, T., Kanda, Y., Sakamaki, Y., Okazaki, M., Li, D., Mizushima, N.
- Source
- Full text @ Elife
|
Loss of PHENOTYPE:
|
|
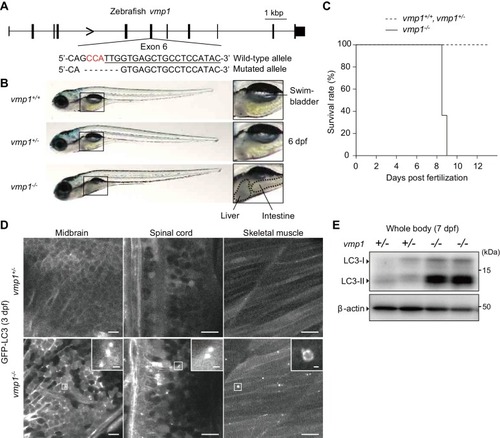
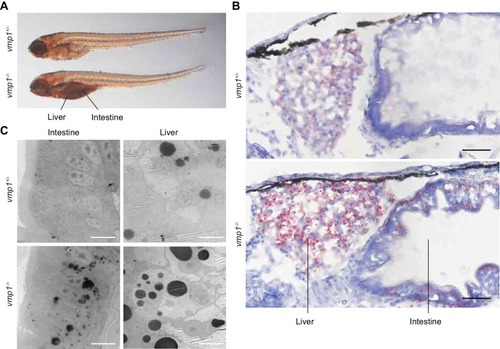
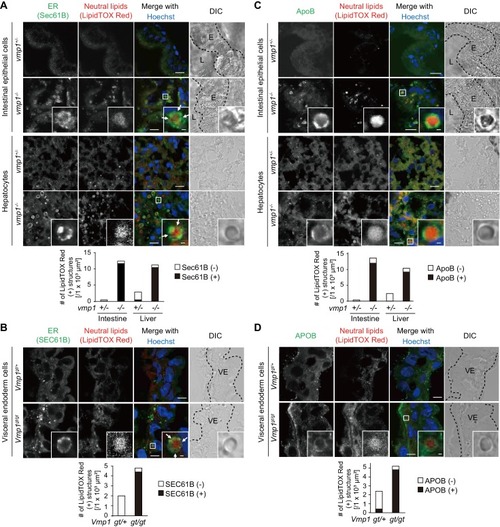
Loss of vmp1 in zebrafish causes accumulation of neutral lipids in the intestine and liver. ( |
|
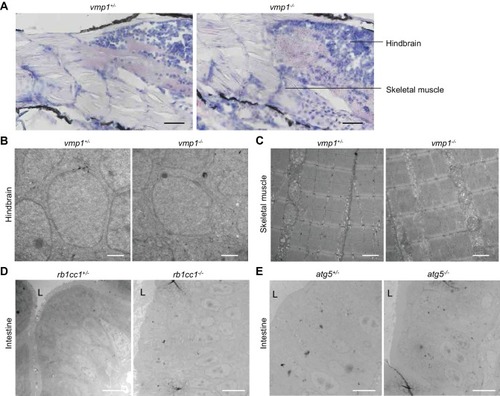
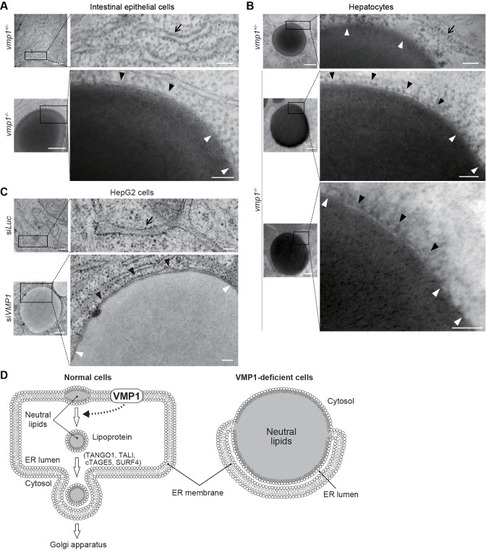
Large lipid-containing structures are not observed in the brain and skeletal muscle of ( PHENOTYPE:
|
|
( |

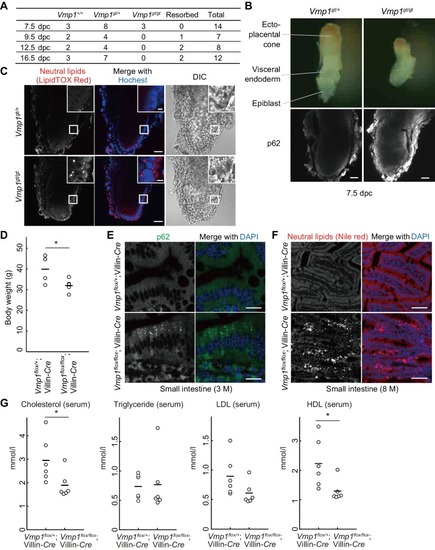
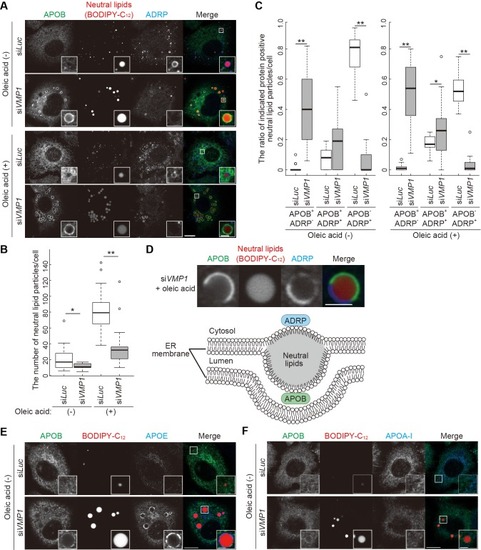
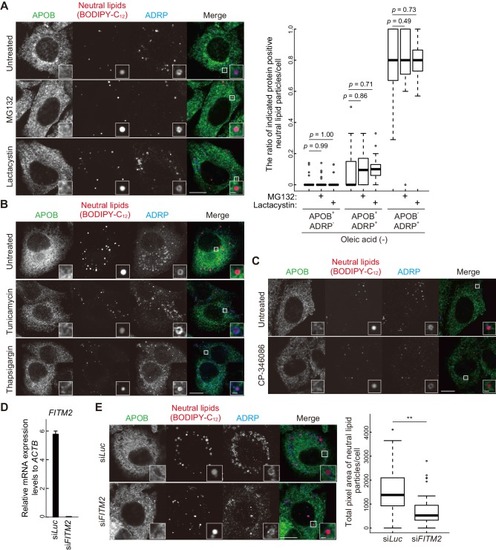
VMP1 is required for secretion and homeostasis of lipoproteins but not for formation of cartilage structures in the zebrafish head skeleton. ( PHENOTYPE:
|
|
PHENOTYPE:
|
|
( |
|
( |
|
( |