- Title
-
Multiplane calcium imaging reveals disrupted development of network topology in zebrafish pcdh19 mutants
- Authors
- Light, S.E.W., Jontes, J.D.
- Source
- Full text @ eNeuro
|
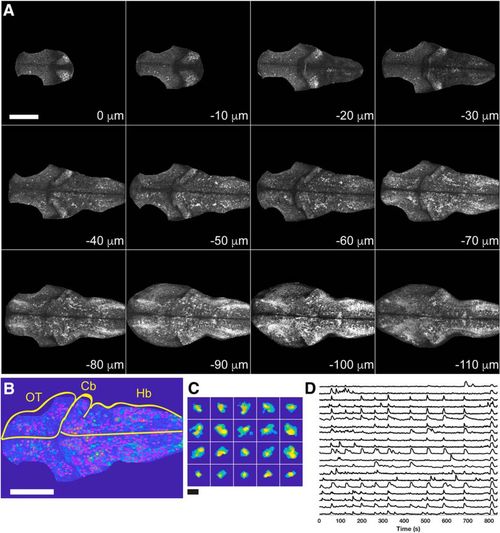
Two-photon imaging of GCaMP6s in zebrafish larvae. A, Each panel shows an optical section from a single time point of a movie collected in a 4 dpf zebrafish larva. Sections are arranged from dorsal (0 μm) to ventral (–110 μm). The brain was cropped to remove skin fluorescence and signal from the synaptic neuropil of the optic tectum. Scale bar = 100 μm. B, Detection of cells identified by constrained non-negative matrix factorization (Pnevmatikakis et al., 2016). ROIs outlined in magenta on top of an average image for a single imaging plane from a 4 dpf larva. Cb, cerebelleum; Hb, hindbrain; OT, optic tectum. Scale bar = 100 μm. C, Selected ROIs. Scale bar = 15 μm. D, Selected ΔF/F traces obtained from the segmented plane in B, C. |
|
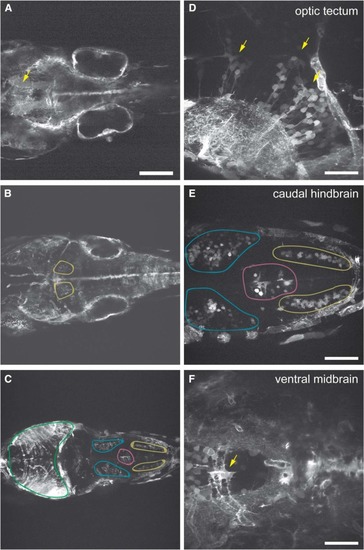
Expression of pcdh19 in BAC transgenic line. |
|
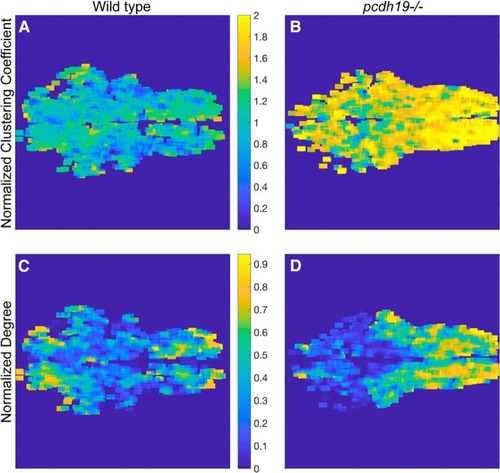
Spatial distribution of node-based measures in a 6 dpf larva. |