- Title
-
The Reprimo Gene Family: A Novel Gene Lineage in Gastric Cancer with Tumor Suppressive Properties
- Authors
- Amigo, J.D., Opazo, J.C., Jorquera, R., Wichmann, I.A., Garcia-Bloj, B.A., Alarcon, M.A., Owen, G.I., Corvalán, A.H.
- Source
- Full text @ Int. J. Mol. Sci.
|
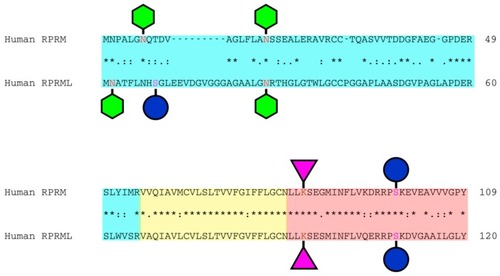
Domain structure and potential post-translational modification sites of human RPRM and RPRML proteins. Schematic representation shows the RPRM and RPRML putative |
|
Domain structure and potential post-translational modification sites of human RPRM and RPRML proteins. Schematic representation shows the RPRM and RPRML putative |
|
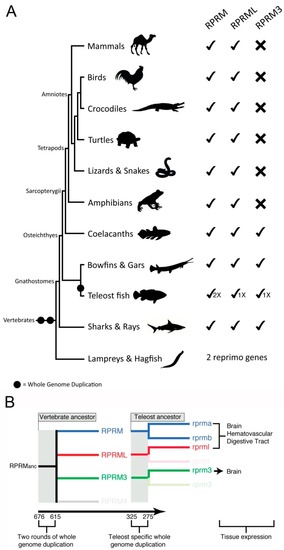
Evolution and diversification of |
|
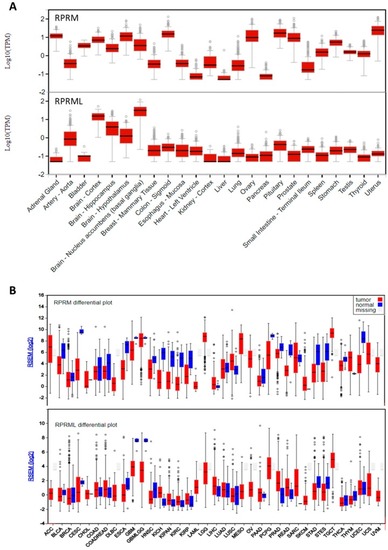
RNA expression of |
|
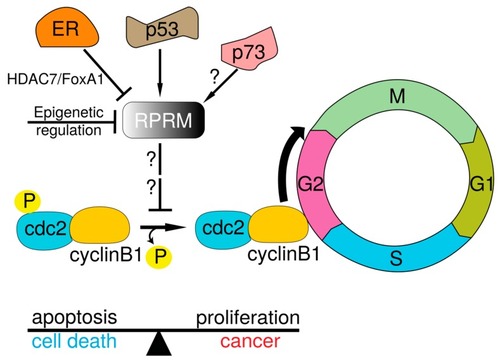
Schematic model of RPRM-mediated cell cycle and G2 arrest mechanisms. |
|
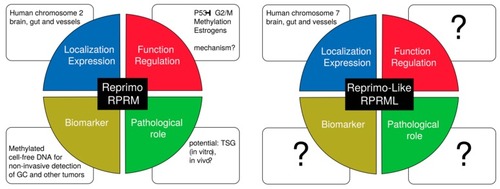
Unanswered questions in |