- Title
-
The Nkd EF-Hand Domain Modulates Divergent Wnt Signaling Outputs in Zebrafish
- Authors
- Marsden, A.N., Derry, S.W., Schneider, I., Scott, C.A., Westfall, T.A., Brastrom, L.K., Shea, M., Dawson, D.V., Slusarski, D.C.
- Source
- Full text @ Dev. Biol.
|
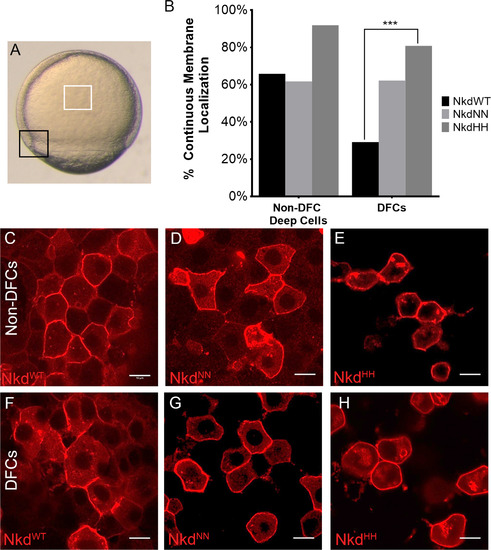
Nkd localization changes in calcium fluxing and quiescent cells. (A) Calcium-fluxing dorsal forerunner cells (DFCs) (black box) and calcium-quiescent non-DFC deep cells (white box) used for localization studies. (B) Analysis of continuous versus discontinuous membrane localization of all Nkd forms in the non-DFC deep cells and the DFCs. NkdWT shows a different localization pattern than NkdNN and NkdHH, which remain mostly membrane-bound. (C and F) NkdWT, NkdNN (D and G), and NkdHH (E and H) localization in the non-DFCs (C-E) and the DFCs (F-H). (F) Quantification of membrane localization. NkdWT shows a significantly different change in localization. ****= P-value<1×10−5, Fisher's exact test, two tail. NkdWT non-DFC n=120, NkdNN non-DFC n=94, NkdHH non-DFCs n=62, NkdWT DFC n=106, NkdNN DFC n=106, NkdHH n=52, n= number of cells. Scale bar 10 µm. Taken at 63x at 2x zoom. See also Fig. S3. |
|
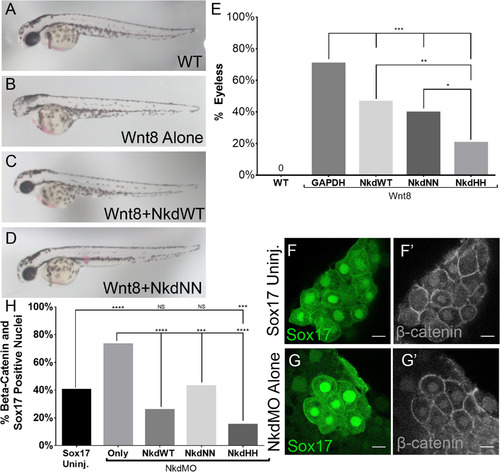
NkdWT, NkdNN, and NkdHHantagonize Wnt/β-catenin. (A-D) Live images of 28hpf embryos, lateral view, anterior to the left. (A) Wild-type, (B) Wnt8+GAPDH RNAs, (C) Wnt8+NkdWT RNAs, and (D) Wnt8+NkdNN RNAs. (E) Graph of percentage of embryos without eyes. hpf, hours post fertilization. Uninj n=230, Wnt8 alone n=148, Wnt8+GAPDH n=34, Wnt8+NkdWT n=71, Wnt8+NkdNN n=44, Wnt8+NkdHH n=53. ***=p-value<0.0001, **=p-value<0.001, Fishers Exact Test. (F-G’) Confocal images of DFCs stained with anti- β-catenin. Nkd Antagonizes β-catenin Nuclear Localization in the DFCs. (F and G) Sox17-GFP which marks the DFCs. (F’ and G’) anti-β-catenin staining. (F-F’) Uninjected Sox17. (G-G’) NkdMO injected transgenic Sox17 line. (H) Graph showing percent (%) of embryos with β-catenin and Sox17 positive nuclei. Note that all forms of Nkd were able to rescue morphantsto uninjected levels, and that NkdHH was the most robust. Sox17 uninj. n=211, NkdMO alone n=134, NkdMO+GAPDH n=153, NkdMO+NkdWT n=70, NkdMO+NkdNN n=77, NkdMO+NkdHH n=99. n=number of cells. ****=p-value<1×10−5, ***=p-value<0.0001, Fishers Exact Test. Scale bar 10 µm. Confocal images taken at 63x at 2x zoom. See also Fig. S5. |
|
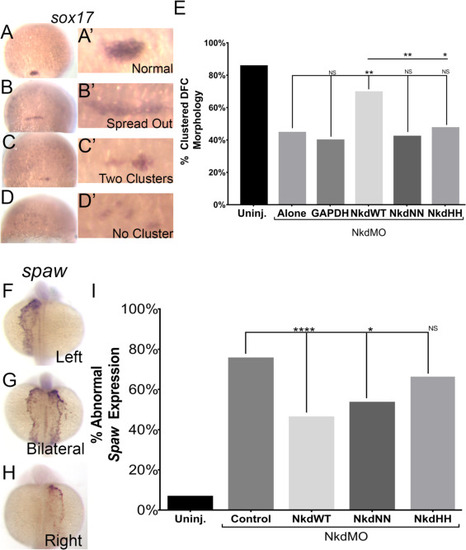
NkdWT, NkdNN, and NkdHHdifferential rescue in Wnt/PCP. (A-E) NkdWT can rescue DFC clusters (A-D’) Images of DFCs from a sox17 in situ, zoom in in A’-D’. (A-A’) Normal, clustered DFC, (B-B’) An abnormal example of spread out DFCs, (C-C’) An abnormal example of two clusters, (D-D’) An abnormal example of a loss of the DFC cluster, (E) NkdWTRNA restores the DFC cluster in NkdMO+NkdWT injected embryos, while NkdNN RNA and NkdHH RNA fails to rescue the cluster morphology of DFCs in NkdMO injected embryos. Uninjected n=145, NkdMO alone n=90, NkdMO+GAPDH n=58, NkdMO+NkdWT n=59, NkdMO+NkdNN n=50, NkdMO+NkdHH n=74. *=p-value<0.01, **=p-value<0.001, ***=p-value<0.0001, Fisher's exact test. (F-I) Nkd Rescue of Left-Right Patterning. (F-H) Spaw in situs on 19–23 ss (18–20 hpf). (F) Normal left-sided expression. (G) Abnormal bilateral expression. (H) Abnormal right-sided expression. (I) NkdWT and NkdNN can rescue abnormal spaw expression, while NkdHH does not. Uninjected n=167, NkdMO+control n=126, NkdMO+NkdWT n=167, NkdMO+NkdNN n=105, NkdMO+NkdHH n=59. Fisher's exact test. *=p-value<0.01, **=p-value<0.001. See also Figs. S6 and S7. |
Reprinted from Developmental Biology, 434(1), Marsden, A.N., Derry, S.W., Schneider, I., Scott, C.A., Westfall, T.A., Brastrom, L.K., Shea, M., Dawson, D.V., Slusarski, D.C., The Nkd EF-Hand Domain Modulates Divergent Wnt Signaling Outputs in Zebrafish, 63-73, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.