- Title
-
Klf8 regulates left-right asymmetric patterning through modulation of Kupffer's vesicle morphogenesis and spaw expression
- Authors
- Lin, C.Y., Tsai, M.Y., Liu, Y.H., Lu, Y.F., Chen, Y.C., Lai, Y.R., Liao, H.C., Lien, H.W., Yang, C.H., Huang, C.J., Hwang, S.L.
- Source
- Full text @ J. Biomed. Sci.
|
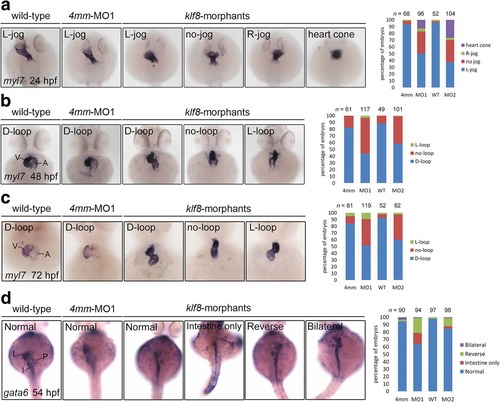
Knockdown of zebrafish klf8 caused defects in heart jogging and looping, and visceral organ positions. a klf8-MO1atg or klf8-MO2atg -injected embryos stained with myl7 exhibited left (L)-jog, no-jog or right (R)-jog and were compared to stained wild-type or 4 mm-MO1-injected control embryos at 24 hpf. b myl7 stained embryos injected with klf8-MO1atg or klf8-MO2atg displayed D-loop, no-loop or L-loop heart and were compared to wild type and control embryos at 48 hpf. c myl7 stained embryos injected with klf8-MO1atg or klf8-MO2atg displayed D-loop, no-loop or L-loop heart and were compared to wild type and control embryos at 72 hpf. d At 54 hpf, gata6 stained wild type or embryos injected with klf8-4 mm MO1, klf8-MO1atg or klf8-MO2atg exhibited organ positions that were classified as: (Normal) normal positions of liver-left, pancreas-right and intestine with left looping, (Intestine only) only intestine without left looping, (Reverse) reversed position of liver-right, pancreas-left and no looping intestine, or (Bilateral) bilateral extension of liver and pancreas. A, atrium; I, intestine; L, liver; P, pancreas; V, ventricle EXPRESSION / LABELING:
PHENOTYPE:
|
|
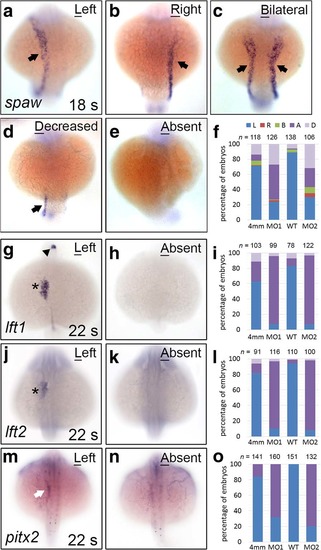
Genes encoding Nodal signalling components exhibited decreased or abolished expression in klf8 morphants. The majority of klf8-MO1atg or klf8-MO2atg -injected embryos showed decreased or abolished spaw (arrow) expression in the left lateral plate mesoderm (LPM) at the 18 s stage (a-f). The majority of klf8 morphants failed to express lft1 in the left diencephalon (arrowhead) or heart (asterisk) (g- i), lft2 in the left heart (asterisk) (j-l), and pitx2 in the left LPM (white arrow) (m-o) at the 22 s stage. Dorsal views of embryos are shown. A, absent; B, bilateral; D, decreased; L, left; R, right EXPRESSION / LABELING:
PHENOTYPE:
|
|
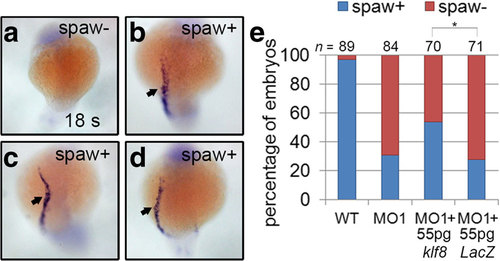
Reduced spaw expression in klf8 morphants was partially rescued by co-injection of klf8 mRNA. Representative embryos showing spaw expression in the left LPM (spaw+) or absent spaw expression (spaw-) are shown (a-d). Percentages of embryos with asymmetric spaw expression or no spaw expression are shown with indicated treatments (e). Statistical significance was determined by Fisher’s Exact Test. *p < 0.05 EXPRESSION / LABELING:
PHENOTYPE:
|
|
Overexpression of klf8 mRNA caused bilateral expression of Nodal signalling component genes. Injection of klf8 mRNA induced bilateral expression of spaw in the LPM (arrow) at the 18 s stage, in a dose-dependent manner (a-e). Embryos injected with klf8, but not LacZ, exhibited bilateral expression of lft1 in the diencephalon (arrowhead) and heart (asterisk; f-j), lft2 in the heart (asterisk; k-o), and pitx2 in the LPM (white arrow) (p-t) at the 19–22 s stage. Expression of ntl in the notochord is similar in wild type and embryos injected with LacZ or klf8 mRNA (u-w). A, absent; B, bilateral; L, left; R, right |
|
Knockdown of klf8 affected dorsal forerunner cell number, KV cilia number and length. Cell number of dorsal forerunner cells (DFCs) was affected in klf8-MO1atg- or klf8-MO2atg -injected embryos as compared to klf8-4 mm MO1-injected Tg(sox17:gfp) control embryos (4 mm) at 75% epiboly (a-d). Images of KV cilia stained with acetylated tubulin antibody in klf8 morphants (g-i) and control embryos (e, f) at 10s stage (e-i). Images of acetylated tubulin stained KV cilia and GFP stained DFCs in Tg(sox17:gfp) embryos injected with different klf8 MOs (l-n), klf8-4 mm MO1 (k) or wild type (j) embryos at 10s stage (j-n). DFC number (o), KV cilia number (q), length (r) but not lumen area (p) were affected in klf8 morphants as compared to control embryos. Statistical significance was determined by Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars indicate standard deviation EXPRESSION / LABELING:
PHENOTYPE:
|
|
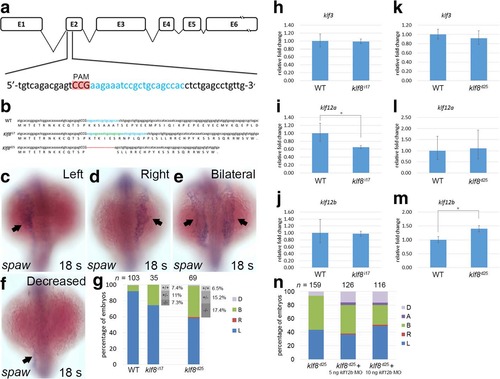
Generation of klf8 mutants by CRISPR-Cas9 gene editing and the effect on spaw expression. a klf8 genomic structure with klf8 sgRNA (blue lettering) targeted to exon 2. Protospacer adjacent motif (PAM) sequence is shown in red. b Nucleotide and predicted amino acid sequences of klf8 in wild type, klf8 I17 and klf8d25 mutants are shown. Deleted nucleotides are shown by a red dashed line, while inserted nucleotides are shown in green lettering. Representative images of embryos with different spaw expression patterns in the LPM at 18 s stage (arrow; c-f). g Percentage of embryos displayed left (L), right (R), decreased (D) or bilateral (B) expression of spaw in the LPM from intercross of respective klf8 d25 or klf8 i17 F2 heterozygous mutants. Deduced percentage of wild type (+/+), heterozygote (+/−) or homozygote (−/−) genotype of embryos from intercross of respective klf8 d25 or klf8 i17 F2 heterozygous mutants exhibited bilateral spaw expression pattern. Expression levels of klf3 (h, k), klf12a (i, l) or klf12b (j, m) were compared between wild type and respective klf8 d25 and klf8 i17 F5 homozygous mutant embryos at 10–12 s stages (h-m). Knockdown of klf12b reduced the percentage of embryos with bilateral spaw expression in the LPM of klf8 d25 F6 homozygous mutant embryos, but the reduction did not reach significance (p = 0.45 for the comparison between klf8 d25 and klf8 d25 + 5 ng klf812b MO, p = 0.05 for the comparison between klf8 d25 and klf8 d25 + 10 ng klf812b MO) (n). Statistical significance was determined by Student’s t-test. * p < 0.05. Error bars indicate standard deviation EXPRESSION / LABELING:
PHENOTYPE:
|
|
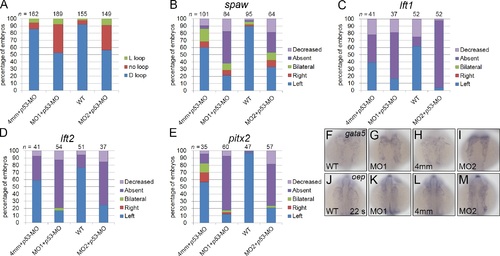
Heart looping and downregulated expression of spaw and its downstream genes were not caused by induction of p53 expression in klf8 morphants. Embryos co-injected with p53-MOsp and klf8-MO1atg or klf8-MO2atg displayed no-loop or L-loop heart defects at 72 hpf (A). The majority of embryos co-injected with p53-MOsp and klf8-MO1atg or klf8-MO2atg exhibited decreased or absent expression of spaw (B), lft1 (C), lft2 (D), or pitx2 (E) in the left LPM, diencephalon or heart at the 18–22 s stages. Expression levels of gata5 and oep which are known to be expressed in the LPM at the 22 s stage were unaffected by klf8 knockdown (F-M). |
|
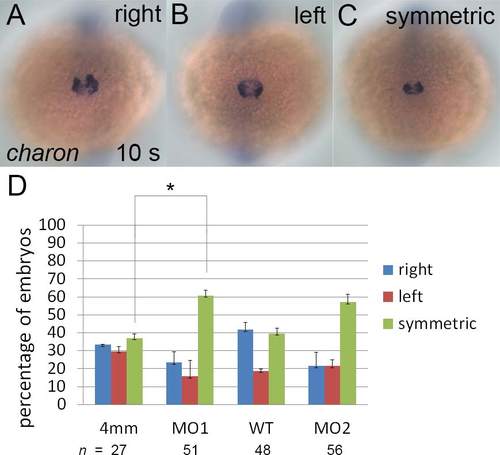
Symmetric charon expression around KV was observed in the majority of klf8 morphants. Representative images of embryos showing stronger charon expression on the right side (A) or left side (B) and symmetric charon expression on both sides (C) of KV are shown. Quantification of different charon expression patterns in embryos injected with different klf8 MOs, control MO or wild type embryo is shown (D). Statistical significance was determined by Student’s t-test. * p < 0.05. Error bars indicate standard deviation. |
|
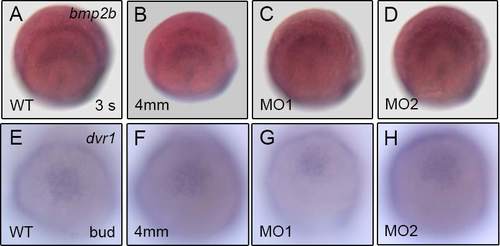
Expression level of bmp2b or dvr1 around tailbud region was not affected by klf8 knockdown. Representative images show similar expression level of bmp2b (A-D) or dvr1 (E-H) around the tailbud region in the wild type or embryos injected with klf8-MO1atg, klf8-MO2atg or klf8-4 mm MO1 at 3 s or bud stages. |