- Title
-
Tol2 gene trap integrations in the zebrafish amyloid precursor protein genes appa and aplp2 reveal accumulation of secreted APP at the embryonic veins
- Authors
- Liao, H.K., Wang, Y., Watt, K.E., Wen, Q., Breitback, J., Kemmet, C.K., Clark, K.J., Ekker, S.C., Essner, J.J., and McGrail, M.
- Source
- Full text @ Dev. Dyn.
|
Phylogenetic and expression analysis of zebrafish appa and aplp2. A: Phylogenetic tree of amyloid precursor proteins. Zebrafish Appa and Aplp2 are shown in bold. The horizontal bars represent the percentage of amino acid substitutions required to generate the corresponding tree. Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Xl, Xenopus laevis. B: Relative expression levels of appa and aplp2 throughout zebrafish development and in adult tissues as evaluated by reverse transcriptase-polymerase chain reaction. Expression of the ribosomal protein S6 kinase b, polypeptide 1b (rps6kb1) gene was used as a control. |
|
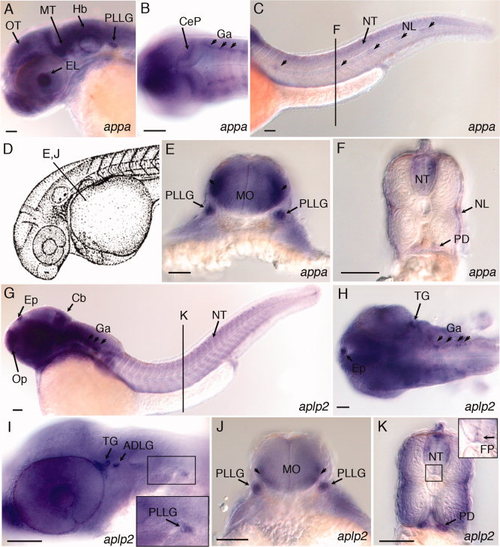
Expression of appa and aplp2 in developing central and peripheral nervous system of 36 hpf zebrafish embryos. A: Lateral view, appa expression in the eye lens, optic tectum, midbrain tegmentum, hindbrain, and posterior lateral line ganglion. B: Dorsal view, appa expression in the cerebellar plate and ganglia. C: Lateral view, appa expression in the neural tube and lateral line neuromasts (arrowheads). Line labeled “F” refers to position of cross-section shown in panel F. D: Camera lucida drawing of 35 hours post fertilization (hpf) zebrafish embryo (Kimmel et al.,1995). Line labeled “E, J” refers to position of cross-sections in panels E and J. E:appa expression in the posterior lateral line ganglia and lateral regions of medulla oblongata (arrowheads). F:appa expression in the neural tube, neuromasts of the lateral line, and pronephric ducts. G: Lateral view of aplp2 expression in the epiphysis, olfactory placode, cerebellum, ganglia, and neural tube. Line labeled “K” refers to position of cross-section shown in panel K. H: Dorsal view, aplp2 expression in the epiphysis, trigeminal ganglia, and ganglia in the hindbrain. I: Lateral view, aplp2 expression in the trigeminal ganglia, anterodorsal and posterior lateral line ganglia. J:aplp2 expression in posterior lateral line ganglion and lateral regions of medulla oblongata (arrowheads). K:aplp2 expression in lateral regions of the neural tube, floor plate, and pronephric duct. Inset shows higher magnification view of boxed region of neural tube. Arrow points to expression of aplp2 in the floor plate region. ADLG, anterodorsal lateral line ganglia; Cb, cerebellum; CeP, cerebellar plate; EL, eye lens; Ep, epiphysis; FP, floor plate; Ga, ganglion; Hb, Hindbrain; MO, medulla oblongata; MT, midbrain tegmentum; NL, neuromasts of lateral line; NT, neural tube; Op, olfactory placode; OT, optic tectum; PD, pronephric duct; PLLG, posterior lateral line ganglion; TG, trigeminal ganglia. Scale bars = 50 μm. EXPRESSION / LABELING:
|
|
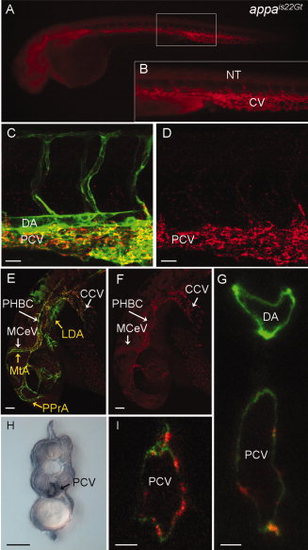
Red fluorescent protein (RFP) expression in the appais22Gt gene trap line. A–G,I: Fluorescence images of live Appais22Gt; Tg(flk1:moesin-gfp)is1 embryos. A: Appa-RFP was detected throughout the veins and weakly in CNS and neural tube in 34 hours post fertilization (hpf) embryos. B: Enlargement of boxed area in (A) shows RFP fluorescence in the caudal vein and intersegmental vessels. C–F: Appa-RFP localization at 32 hpf embryos in the trunk (C,D) and the head (E,F). Appa-RFP fluorescence overlaps with GFP expression in endothelial cells in the veins (C,E), but not the arterial vessels (MTA, PPrA, LDA marked with yellow arrows in E). H: Immunolocalization with anti-RFP antibody shows Appa-RFP is localized to the caudal vein but absent from the dorsal aorta. G,I: Confocal images of cross-section through the trunk of a 36 hpf embryo shows Appa-RFP accumulation in the posterior caudal vein. CCV, common cardinal vein; CV, caudal vein; LDA, lateral dorsal aorta; MCeV, midcerebral vein; MtA, metencephalic artery; NT, neural tube; PHBC, primordial hindbrain channel; PPrA, primitive prosencephalic artery. Scale bars = 20 μm in C,D, 50 μm in E–I. |
|
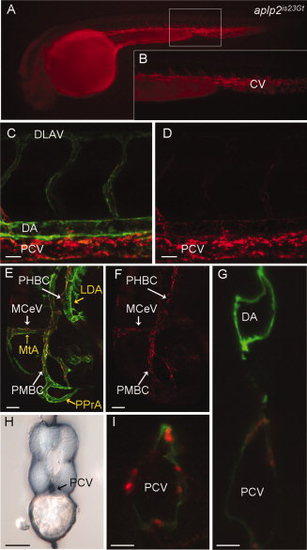
Red fluorescent protein (RFP) expression in the aplp2is23Gt gene trap line. Fluorescence images of live aplp2is23Gt; Tg(flk1:moesin-gfp)is1 embryos. A: Aplp2-RFP fluorescence in the caudal vein and intersegmental vessels in a 34 hours post fertilization (hpf) embryo. B: Enlargement of boxed area in (A). C–F: RFP fluorescence at 32 hpf was observed in the trunk (C,D) and the head (E,F). C,E: Aplp2-RFP fluorescence overlaps with green fluorescent protein (GFP) expression in endothelial cells in the veins, but not the arterial vessels (MTA, PPrA, LDA marked with yellow arrows in E). Weak expression is detected in the dorsal aorta. H: Immunolocalization with anti-RFP antibody shows Appa-RFP was in the caudal vein but absent from the dorsal aorta. G,I: Confocal images of cross-sections through the trunk of 36 hpf embryo shows Aplp2-RFP accumulation at the posterior caudal vein. CV, caudal vein; LDA, lateral dorsal aorta; MCeV, midcerebral vein; MtA, metencephalic artery; PHBC, primordial hindbrain channel; PMBC, primordial midbrain channel; PPrA, primitive prosencephalic artery. Scale bars = 20 μm in C,D, 50 μm in E–I. |
|
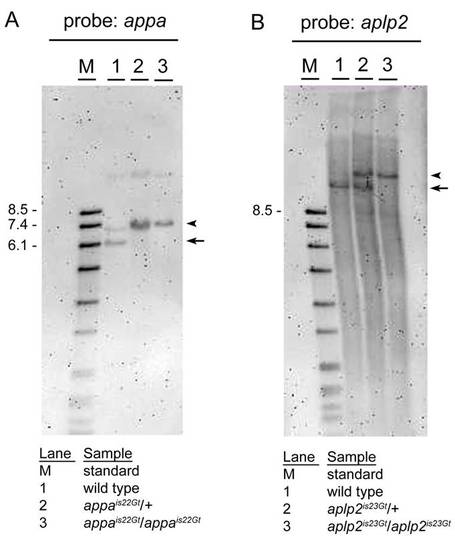
Gene trap lines appais22Gt and aplp2is23Gt contain Tol2 transposon integrations. A: SphI digested genomic DNA from wild-type (1), heterozygous appais22Gt/+ (2), and homozygous appais22Gt/is22Gt (3) adults, blotted and hybridized with an appa specific probe. The wild-type restriction fragment of <6.2 kb (arrow) is shifted to <7.2 kb (arrowhead) as expected due to the Tol2 integration in transgenic appais22Gt embryos. The faint higher band in lane 1 might be due to incomplete digestion or polymorphism in the appa gene in the WIK wild-type strain. The sample run in lane 2 is most likely DNA from a homozygous instead of heterozygous adult. B: SacI digested genomic DNA from wild-type (1), heterozygous aplp2is23Gt/+ (2), and homozygous aplp2is23Gt/is23Gt (3) adults, blotted and hybridized with an aplp2 specific probe. The wild-type restriction fragment of <12.1 kb in the aplp2 gene (arrow) is shifted to the expected size of <15.1 kb (arrowhead), due to the <3 kb Tol2 transposon integration, in the heterozygous aplp2is23Gt/+ adult. The homozygous aplp2is23Gt/is23Gt adult has one band at the expected size of <15.1 kb. |
|
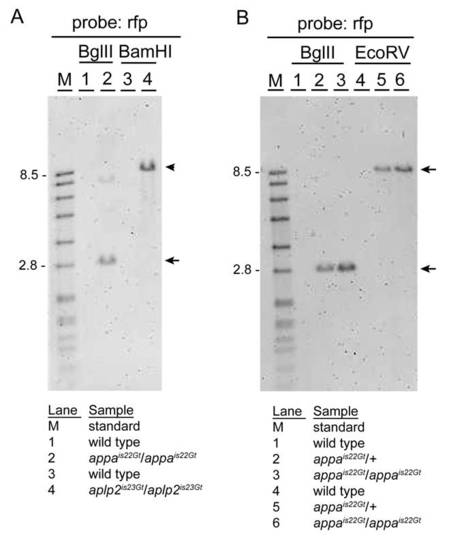
Gene trap lines appais22Gt and aplp2is23Gt contain single Tol2 transposon integrations. A: Genomic Southern blot hybridized with red fluorescent protein (RFP) probe. DNA from wild-type (1) and homozygous appais22Gt/is22Gt adults (2) digested with BglII; wild-type (3) and homozygous aplp2is23Gt/is23Gt adults (4) digested with BamHI. As predicted based on the location of the Tol2 transposon integration site in the appa gene, a single BglII restriction fragment of <2.8 kb (arrow) is present in the appais22Gt/is22Gt adult. The faint higher band is most likely due to incomplete digestion (see B, repeat of BglII digestion). In BamHI digested DNA from the aplp2is23Gt/is23Gt adult, a single restriction fragment of <7.9 kb (arrowhead) is detected. This is the expected size based on the predicted integration site of the Tol2 transposon in the aplp2 gene in line aplp2is23Gt. B: To confirm the presence of a single Tol2 transposon integration in the appa gene in line appais22Gt, genomic DNA from wild-type, heterozygous appais22Gt/+, and homozygous appais22Gt/is22Gt adults was digested with BglII or EcoRV, blotted, and hybridized with the RFP-specific probe. A single restriction fragment of the expected size (2.8 kb BglII; 8.9 kb EcoRV) is detected in heterozygous and homozygous appais22Gt adults (arrow and arrowhead). |
|
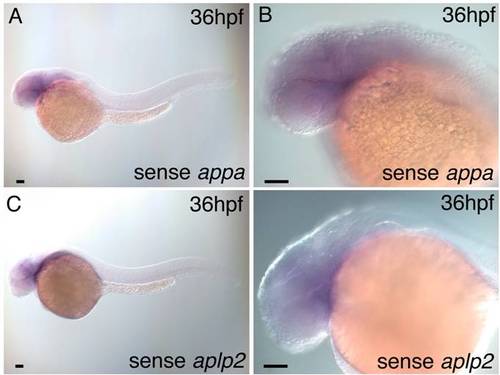
Wild-type embryo in situ hybridization with sense appa and aplp2 probes. A,B: A 36 hours post fertilization (hpf) wild-type embryo hybridized with sense appa probe shows no detectable expression above background. C,D: A 36 hpf wild-type embryo hybridized with sense aplp2 probe shows no detectable expression above background. Scale bars = 50 μm. |
|
rfp mRNA expression in appais22Gt and aplp2is23Gt embryos by in situ hybridization. A: RNA expression in the midbrain, cerebellum, and epiphysis in an appais22Gt embryo. F: Expression of rfp in the neural tube and pronephric ducts of an appais22Gt embryo. Line refers to position in trunk of cross-section shown in insert. Insert, rfp expression is detected in the neural tube, lateral line, and pronephric ducts. B,G: Nontransgenic sibling of the embryos in panels A and F shows no expression above background. C: RNA expression in the midbrain, cerebellum, and epiphysis in an aplp2is23Gt embryo. H: RNA expression in the pronephric ducts of an aplp2is23Gt embryo. Line refers to the position of the cross-section through the trunk shown in inset. D,I: Nontransgenic sibling of embryos shown in C and H. E,J: Wild-type embryos hybridized with rfp probe shown background level of expression. Cb, cerebellum; Ep, epiphsis; NL, neuromasts of lateral line; NT, neural tube; PD, pronephric ducts. Scale bars = 100 μm. |
|
Confocal imaging of live appa+/is22Gt; Tg(flk1:moesin1-egfp)+/is1 embryo before completion of venous vessel formation. A–D: Lateral view, 24 hours post fertilization (hpf). A: Appa-red fluorescent protein (RFP) in the head is enriched in the eye surrounding the lens (yellow arrows). Appa-RFP has not accumulated at the veins in the head. C,D: Appa-RFP accumulates at the somite boundaries in the trunk (yellow arrow) but is absent from the posterior caudal vein region (white arrow). E–H: Lateral view, 28 hpf. E,F: Appa-RFP does not show enrichment at the veins in the head at this time. G,H: Accumulation of Appa-RFP at the somite boundaries has increased (yellow arrows) and is detected in the region of the posterior caudal vein (white arrows). Scale bars = 50 μm. |
|
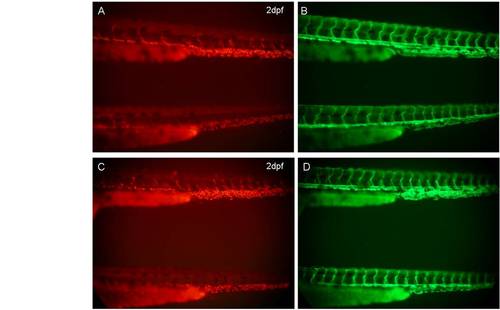
Localization of Appa-red fluorescent protein (RFP) and Aplp2-RFP in the vasculature does not require blood flow. A,B: Lateral views of appa+/is22Gt; Tg(flk1:moesin1-egfp)+/is1; sih+/mn0031Gt (upper) and appa+/is22Gt Tg(flk1:moesin1-egfp)+/is1; sihmn0031Gt/mn0031Gt (bottom) embryos at 2 dpf. C,D: Lateral views of aplp2+/is23Gt; Tg(flk1:moesin1-egfp)+/is1; sih+/mn0031Gt (upper) and aplp2+/is23Gt; Tg(flk1:moesin1-egfp)+/is1; sihmn0031Gt/mn0031Gt (bottom) embryos at 2 dpf. Both Appa-RFP and Aplp2-RFP were detected in the vein region in sih homozygous mutant embryos. |
|
Expression of Appa-red fluorescent protein (RFP) and Aplp2-RFP in appais22Gt and aplp2is23Gt adult tissues. A,B: Appa-RFP localization in cells embedded in the membrane covering the brain. C–E: Appa-RFP localization in kidney tubules (arrows). F–I: Aplp2-RFP localization in cells embedded in the membrane covering the brain. J: Aplp2-RFP localization in kidney tubules (arrows). |
|
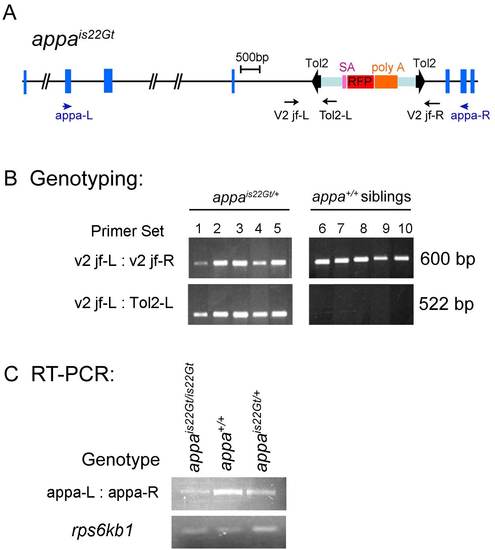
DNA genotyping and reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of wild-type transcript levels in appais22Gt embryos. A: Structure of the appa locus with is22Gt Tol2 trap integration site. Blue boxes indicate exons. The Tol2 gene trap transposon is shown within bold black arrowheads. Purple, red, and orange boxes indicate the splice acceptor, the RFP cDNA, and the transcriptional terminator / polyadenylation sequence (poly A), respectively. Position and direction of primers for genomic DNA genotyping (black) and RT-PCR analysis (blue) are shown as arrows. B: Genomic DNA genotyping of appa heterozygous embryos and wild-type siblings. Control primers v2 jf-L and v2 jf-R amplify a 600-bp fragment of the wild-type appa gene present in wild-type and heterozygous appais22Gt/+ embryos. A 522-bp fragment spanning the junction of the Tol2 transposon integration site in appa amplifies with the v2 jf-L primer and the Tol2-L primer, which sits inside the left inverted repeat of the Tol2 transposon. The 522-bp fragment is present in heterozygous appais22Gt/+ embryos but absent from wild-type siblings. C: RT-PCR analysis on RNA isolated from appais22Gt homozygous, heterozygous and wild-type embryos. The appa-L and appa-R primers used for RT-PCR sit in exons 4 and 9, respectively, of the appa gene. The normally spliced appa mRNA is amplified by RT-PCR in the homozygous appais22Gt/is22Gt embryos, however, the amount is greatly reduced compared with heterozygous and wild-type embryos. Ribosomal protein rps6kb1 primers were used for control. |