- Title
-
A cluster of non-redundant Ngn1 binding sites is required for regulation of deltaA expression in zebrafish
- Authors
- Madelaine, R., and Blader, P.
- Source
- Full text @ Dev. Biol.
|
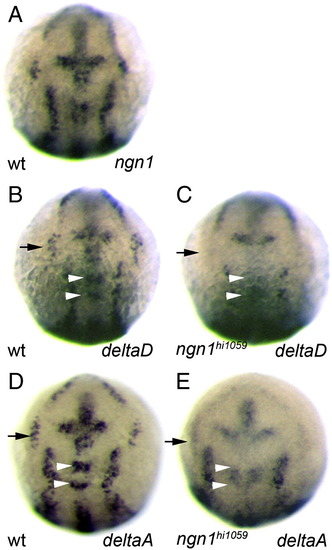
Loss of Ngn1 function leads to loss of deltaA and deltaD expression. Whole-mount in situ hybridization against ngn1 (A), deltaD (B, C) or deltaA (D, E) in zebrafish embryos at 11 hpf. Expression in wild type embryos show that ngn1, deltaD and deltaA share similar expression patterns (A, B, D). In ngn1 mutant embryos (C, E), deltaD and deltaA expression is strongly reduced in the anterior neural plate; this is most notable for the trigeminal ganglia (black arrows in B–E) and the medial clusters in rhombomeres 2 and 4 (white arrowheads in B–E). Embryos are viewed dorsally with anterior up. |
|
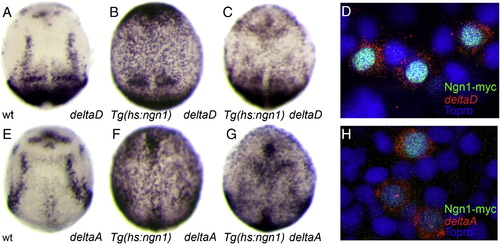
Activation of deltaA and deltaD by Ngn1 is rapid and cell autonomous. Whole-mount in situ hybridization against deltaD (A–D) and deltaA (E–H) in embryos at 9 hpf (C, G) or 10 hpf (A, B, D, E, F, H). Tg(hs:ngn1) embryos heat-shocked at 7 hpf display ectopic expression of deltaD and deltaA 2 h after treatment (B, F). Similarly, mis-expression of Ngn1 at 8.5 hpf induces deltaD and deltaA expression in as little as 30 min (C, G). Single confocal sections of the neural plate at 10 hpf show that mosaic mis-expression of a Myc-tagged form of Ngn1 (in green) induces deltaD and deltaA (in red) in an apparently cell autonomous manner (D, H); nuclei are labelled with Topro (blue) to show that there are cells in the field of view that do not express Ngn1-myc or delta and that induced delta transcripts form a halo around the Ngn1-myc positive nucleus. Embryos are viewed dorsally with anterior up. |
|
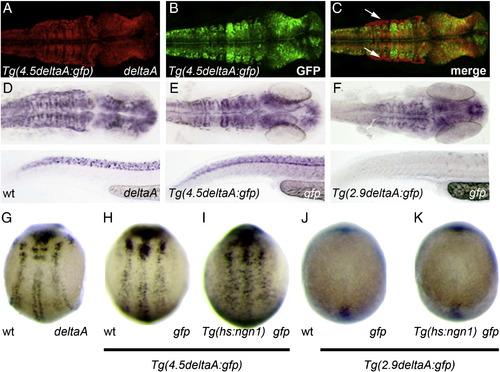
A ngn-regulated cis-regulatory module is located between -2.9 and -4.5 kb upstream of deltaA. (A–C) Confocal projections of double in situ/immunolabelling showing extensive overlap in the expression of endogenous deltaA mRNA and GFP protein expression in Tg(4.5delA:GFP) embryos at 24 hpf; a notable exception is the dorsal limit of the hindbrain that expresses deltaA but where GFP is not detected (white arrows in C). (D–F) Whole-mount in situ hybridization against deltaA (D) or gfp in Tg(4.5delA:GFP) (E) versus Tg(2.9delA:GFP) (F) transgenic embryos at 24 hpf. The shorter transgene drives gfp expression in a significantly more restricted pattern. (G, H) The expression of endogenous deltaA and GFP expression in Tg(4.5delA:GFP) embryos at 10 hpf are highly similar. (H–K) Mis-expression of Ngn1 induces ectopic expression of gfp in 10 hpf Tg(4.5deltaA:GFP) embryos but not in stage-matched Tg(2.9deltaA:GFP) embryos. Embryos at 24 hpf are mounted with anterior to the right; the brain is viewed dorsally and the trunk and tail are viewed laterally with dorsal part up; at 10 hpf, embryos are viewed dorsally with anterior up. |
|
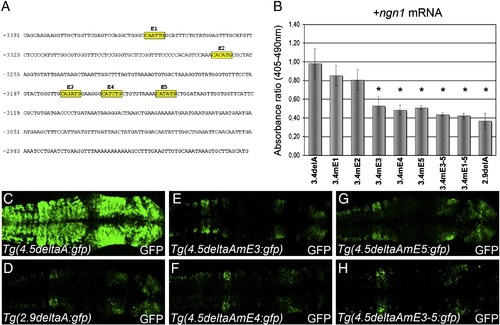
E-boxes are required for the transcriptional regulation of deltaA by Ngn1. (A) Nucleotide sequence of genomic DNA located between -3.4 and -2.9 kb upstream of the DeltaA coding region. Yellow boxes highlight the five E-boxes in this sequence. (B) Histogram showing the ratio of CAT levels after the mis-expression of various promoter:reporter constructs and synthetic ngn1 mRNA. CAT expression levels were normalized against LacZ driven off the 7.1deltaA:LacZ fragment. (C–H) Confocal projections after immunohistochemical detection of GFP showing expression in the anterior neural tube in a series of stable transgenic embryos at 24 hpf. Whereas Tg(4.5deltaA:GFP) embryos show strong and widespread GFP expression (C) the Tg(2.9deltaA:GFP) transgene drives weaker GFP expression in a more restricted pattern (D). Transgenic lines where individual E-box (E–G) or the three clustered E-boxes were mutated (H) display GFP expression similar to Tg(2.9deltaA:GFP). Embryos are viewed dorsally and mounted with anterior to the right. Error bars represent s.d. *P < 0.05; **P < 0.001; ***P < 0.0005 using a t-test. |
|
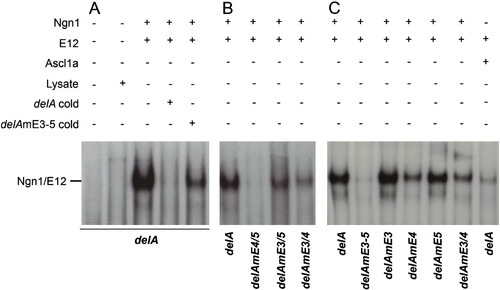
Specific DNA binding of Ngn1/E12 to the deltaA CRM. In vitro transcribed/translated Ngn1, Ascl1a and E12 were assayed by gel mobility shift assay (EMSA) for binding to a delA probe or similar probes containing mutations in specific E-boxes. (A) Ngn1/E12 heterodimers bind to delA and an excess of cold probe efficiently competes this binding. Little competition is detected with cold probes carrying mutations in the three E-boxes. (B) Different affinities of Ngn1/E12 heterodimer binding for individual E-boxes are highlighted by EMSA using delA probes carrying mutations in different pairs of E-boxes; while binding is strong to E4 (delAEm3/5), binding is poor to E5 (delAmE3/4) and virtually undetectable to E3 (delAEm4/5). (C) Different affinities of Ngn1/E12 heterodimer binding to couplets of E-boxes are highlighted by EMSA using delA probes carrying mutations in individual E-boxes. While Ngn1/E12 binding is strong to E4/E5 (delAEm3) and E3/E4 (delAEm5), binding is strongly decreased when E4 is mutated (delAEm4); binding is lost when the three E-boxes are mutated (delAEm3–5) and the wildtype delA probe is bound more specifically by Ngn1/E12 than Ascl1a/E12. |
Reprinted from Developmental Biology, 350(1), Madelaine, R., and Blader, P., A cluster of non-redundant Ngn1 binding sites is required for regulation of deltaA expression in zebrafish, 198-207, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.