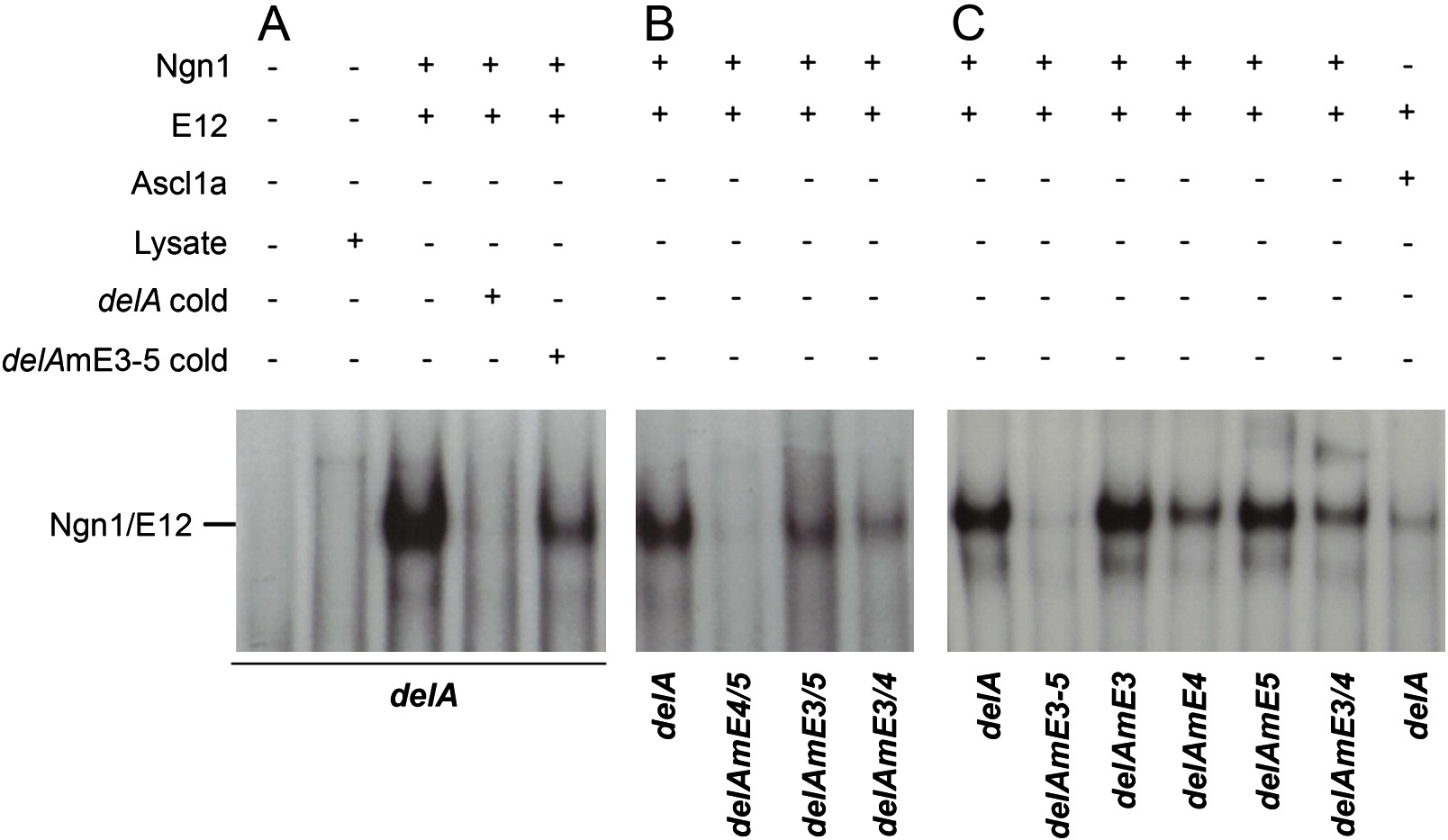
Fig. 6 Specific DNA binding of Ngn1/E12 to the deltaA CRM.
In vitro transcribed/translated Ngn1, Ascl1a and E12 were assayed by gel mobility shift assay (EMSA) for binding to a delA probe or similar probes containing mutations in specific E-boxes. (A) Ngn1/E12 heterodimers bind to delA and an excess of cold probe efficiently competes this binding. Little competition is detected with cold probes carrying mutations in the three E-boxes. (B) Different affinities of Ngn1/E12 heterodimer binding for individual E-boxes are highlighted by EMSA using delA probes carrying mutations in different pairs of E-boxes; while binding is strong to E4 (delAEm3/5), binding is poor to E5 (delAmE3/4) and virtually undetectable to E3 (delAEm4/5). (C) Different affinities of Ngn1/E12 heterodimer binding to couplets of E-boxes are highlighted by EMSA using delA probes carrying mutations in individual E-boxes. While Ngn1/E12 binding is strong to E4/E5 (delAEm3) and E3/E4 (delAEm5), binding is strongly decreased when E4 is mutated (delAEm4); binding is lost when the three E-boxes are mutated (delAEm3–5) and the wildtype delA probe is bound more specifically by Ngn1/E12 than Ascl1a/E12.
Reprinted from Developmental Biology, 350(1), Madelaine, R., and Blader, P., A cluster of non-redundant Ngn1 binding sites is required for regulation of deltaA expression in zebrafish, 198-207, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.