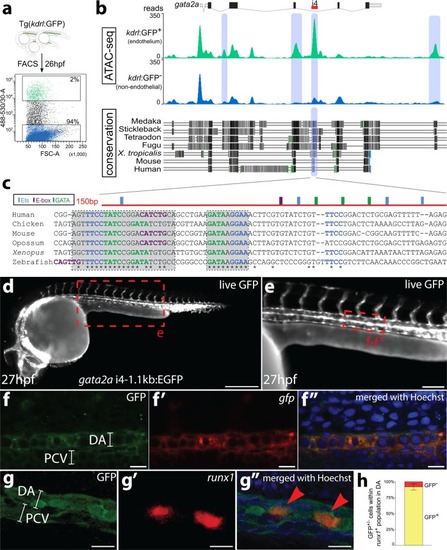
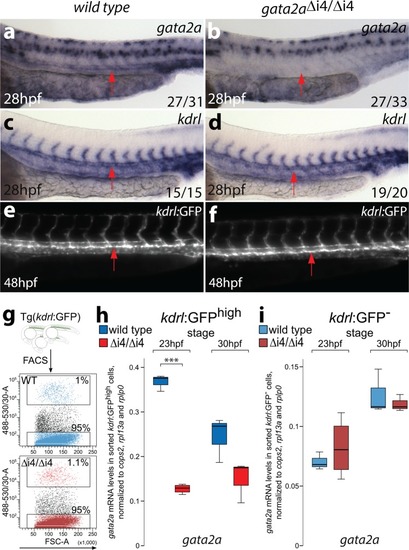
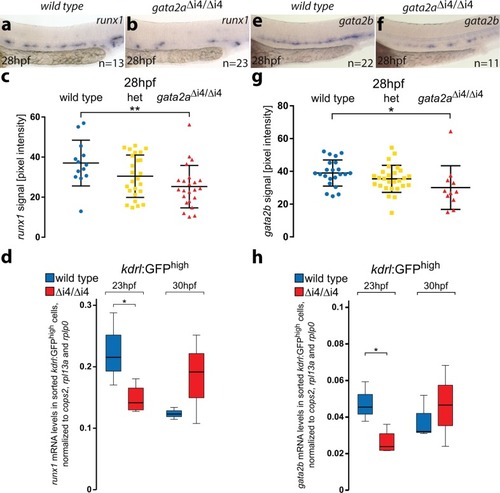
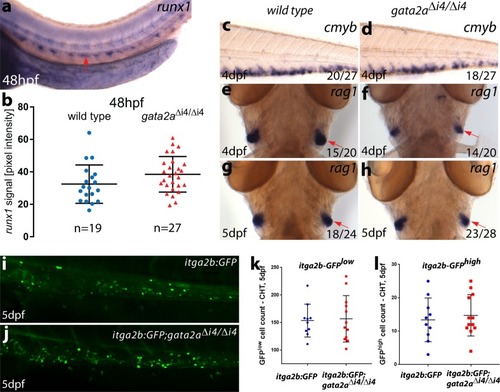
- Title
-
Deletion of a conserved Gata2 enhancer impairs haemogenic endothelium programming and adult Zebrafish haematopoiesis
- Authors
- Dobrzycki, T., Mahony, C.B., Krecsmarik, M., Koyunlar, C., Rispoli, R., Peulen-Zink, J., Gussinklo, K., Fedlaoui, B., de Pater, E., Patient, R., Monteiro, R.
- Source
- Full text @ Commun Biol
|
EXPRESSION / LABELING:
|
|
|
|
|
|
|
|
|
|
PHENOTYPE:
|