- Title
-
CRISPR/Cas9-mediated precise genome modification by a long ssDNA template in zebrafish
- Authors
- Bai, H., Liu, L., An, K., Lu, X., Harrison, M., Zhao, Y., Yan, R., Lu, Z., Li, S., Lin, S., Liang, F., Qin, W.
- Source
- Full text @ BMC Genomics
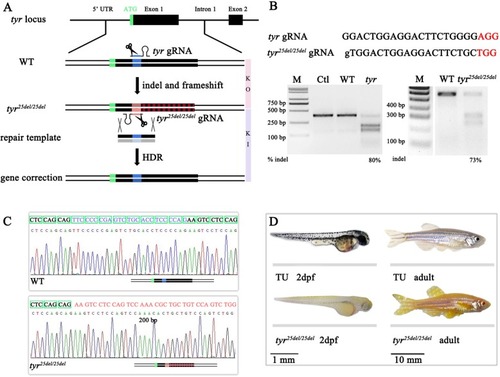
|
CRISPR-mediated |
|
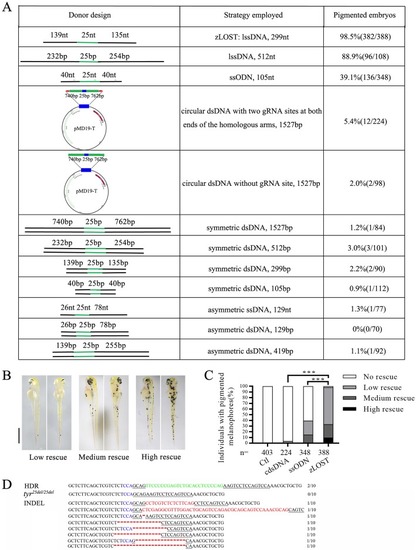
A genetic assay for comparing the efficiency of homology-directed repair using |
|
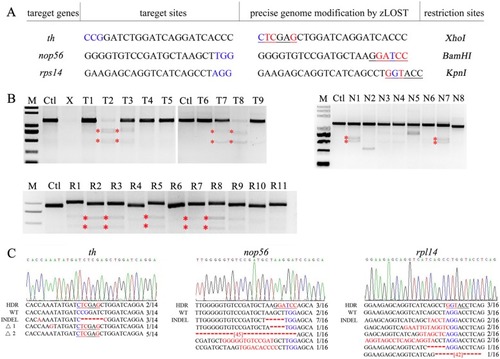
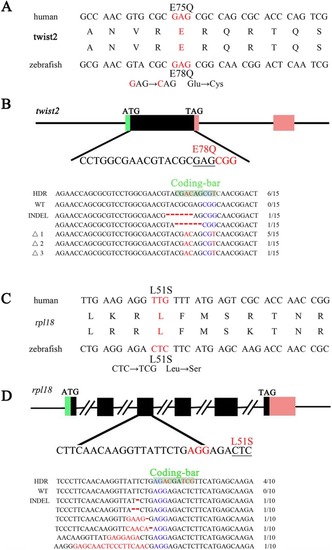
Zebrafish genome editing at three other target sites by zLOST |
|
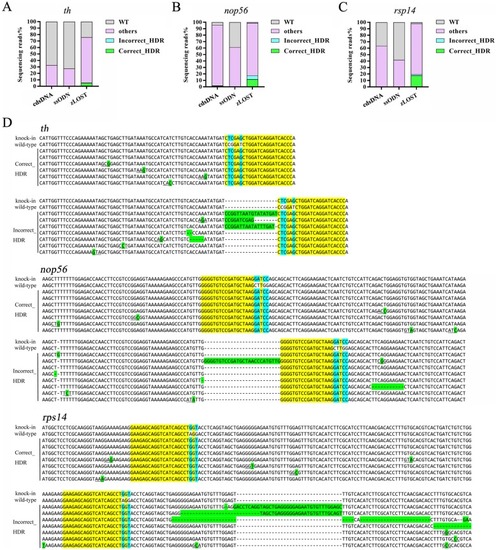
NGS analysis of precise point mutation introduction to the genes Total percentages of defined sequence reads classes at knock-in sites of |
|
zLOST enables mimicking of human disease related mutations in zebrafish Alignment of human patients and desired zebrafish mutations to model human Barber-Say syndrome (BSS) or Diamond-Blackfan anaemia (DBA), schematic outlines of the gene editing strategy and sequencing of the resulting |