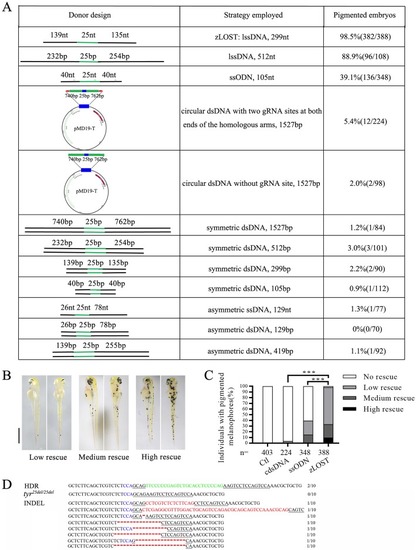
A genetic assay for comparing the efficiency of homology-directed repair using tyr mutant. a Table of template design schematics (left), attributes of the template (middle) and proportion of observed pigmented embryos in the tyr25de/l25del model (right). Embryos are analysed at 2 dpf after co-injection of zCas9 mRNA, tyr25de/l25del gRNA together with repair template. Number of embryos evaluated (n) exceeded 100 for each condition. b Phenotypic evaluation of embryos at 2 dpf into three groups according to number of pigmented cells: low rescue (1–20 pigmented cells), medium rescue (21–40 pigmented cells) and high rescue (more than 40 pigmented cells). Scale bar = 1 mm. c Statistics of HDR efficiency induced by different repair templates. zLOST: long single stranded template 299 bp, ssODN: single strand DNA oligonucleotides 105 bp, cdsDNA: circular double stranded DNAs 1527 bp (with two gRNA sites at both ends of the homologous arms), Ctl: without repair template. Number of embryos assessed (n) is shown for each group. X2-test (***p < 0.001). d Sequence analysis confirming that the larvae contained a correctly repaired tyr locus by zLOST. Correct insertion by HDR (green), PAM region (blue), target sites (underlined), Indels (red) are indicated.
|

