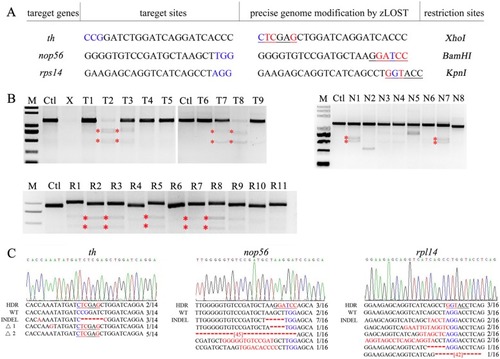
Fig. 3
- ID
- ZDB-FIG-200130-3
- Publication
- Bai et al., 2020 - CRISPR/Cas9-mediated precise genome modification by a long ssDNA template in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
Zebrafish genome editing at three other target sites by zLOST |