- Title
-
Antisense Oligonucleotide-Mediated Transcript Knockdown in Zebrafish
- Authors
- Pauli, A., Montague, T.G., Lennox, K.A., Behlke, M.A., Schier, A.F.
- Source
- Full text @ PLoS One
|
Efficiency and specificity of oep ASO-induced oep (one-eyed-pinhead) mutant phenotypes. A) Injection of oep ASO induces dosage-dependent oep phenotypes that resemble zygotic (Zoep) and maternal-zygotic (MZoep) oep genetic mutants. B) oep ASO and MALAT1 ASO knockdowns are specific. The RNA levels of oep and MALAT1 were measured by qPCR in oep ASO (100 pg) and MALAT1 ASO (80 pg)-injected embryos. Shown is the fold change in RNA level compared to WT (wildtype), normalized to ef1a (error bars: standard deviation of the mean of 3 independent experiments). C) qPCR-based measurement of oep RNA levels in individual oep ASO (100 pg)-injected (red) or uninjected (black) embryos at shield stage (6 hpf). D) Rescue of oep ASO-induced oep phenotypes by coinjection of an oep ASO-resistant RFP-oep fusion mRNA. Note that the oep ASO-sensitive RFP-oep fusion mRNA is efficiently knocked down (no red fluorescence) and does not rescue. E) Quantitation of survival at 24 hpf and F) quantitation of phenotypic strength in survivors at 24 hpf in the presence versus absence of p53 (p53 MO-injected embryos) or RFP-oep fusion mRNA rescue construct. The number of embryos in each category is indicated. |
|
ASO-mediated RNA knockdown correlates with phenotype. A) The RNA level of the gene corresponding to each ASO was measured (compared to WT, normalized to ef1a) and correlated to the presence of a phenotype: green = reproduced published phenotype (in case of MALAT1: no mutant phenotype), orange = reproduced published phenotype in a smaller percentage of embryos, red = did not produce a phenotype. Injected amount of ASO per embryo: 50 pg smad2 ASO, 50 pg smad5 ASO, 50 pg alk8 ASO, 50 pg bmp7a ASO, 150 pg chordin ASO, 100 pg ntla ASO, 50 pg wnt11 ASO, 150 pg tolloid ASO, 100 pg wnt5b ASO, 100 pg nacre ASO, 150 pg MALAT1 ASO. Error bars show standard deviation of the mean of 2 independent experiments (10 embryos each). B) ASO-generated phenotypes for alk8, bmp7a, chordin, smad5 and ctnnb2 (shown are 3 embryos representative of the different severities of ctnnb2 ASO-induced phenotypes). C) ASOs only target the cognate RNA, and not unrelated RNAs. qPCR-based assessment of ASO specificity to their cognate target genes (10 embryos each). D) dnd ASOs block germ cell formation. Germ cells were labeled by injection of 80 pg eGFP-nanos3’UTR mRNA. Coinjection of 25 pg dnd ASO caused complete loss of green germ cells (white arrow). |
|
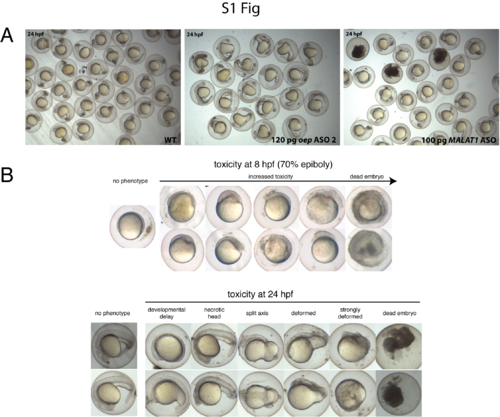
Specificity and toxicity of ASO-mediated RNA knockdown in zebrafish embryos. A) oep ASO-mediated oep mRNA knockdown induces oep-specific phenotypes with high efficiency, while MALAT1 ASO-mediated MALAT1 lncRNA knockdown does not induce any visible, gene-specific phenotypes (3 dead (= black) embryos). B) ASOs induce toxicity at higher concentrations. The concentration at which a specific ASO is toxic for an embryo varies and is ASO sequence-dependent. In general, injection of > 200 pg of an ASO results in general toxicity. Shown are representative images of ASO-injected healthy (= no phenotype), deformed and dead embryos during mid-gastrulation (70% epiboly) and at 24 hpf. |
|
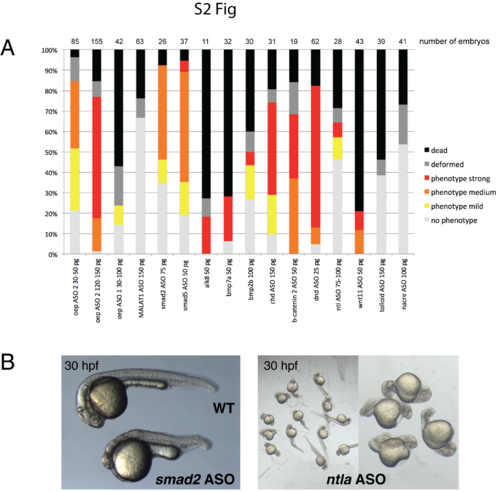
ASO-mediated RNA knockdown is applicable to multiple different transcripts. A) Summary quantitation of survival and phenotypes of ASO-injected embryos at 24 hpf. Percentage plots are derived from multiple independent experiments (at least 2 independent experiments per ASO). Compiled numbers of embryos scored for each ASO are indicated. B) Representative images of smad2 ASO (left) and ntla ASO (right)-injected embryos at 30 hpf. ntla ASO caused a gene-specific phenotype only in a subset of embryos (left: overview image with phenotypic and non-phenotypic embryos; right: higher magnification view of phenotypic embryos). |