- Title
-
Cannabinoid Receptor-2 Regulates Embryonic Hematopoietic Stem Cell Development via PGE2 and P-selectin Activity
- Authors
- Esain, V., Kwan, W., Carroll, K.J., Cortes, M., Liu, S.Y., Frechette, G.M., Sheward, L.M., Nissim, S., Goessling, W., North, T.E.
- Source
- Full text @ Stem Cells
|
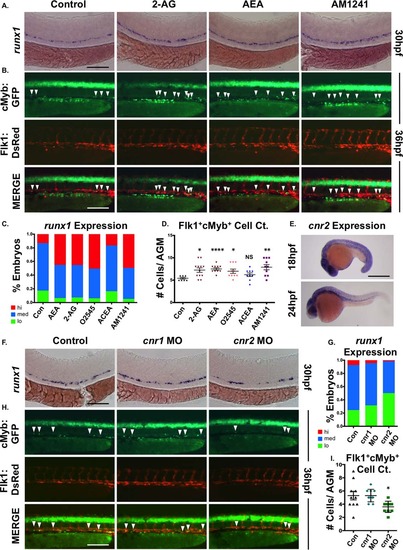
CNR2 modulates hematopoietic stem cell (HSC) formation during embryogenesis. (A): Embryonic exposure to endogenous (2-AG, 5 µM; AEA, 5 µM) or synthetic (AM1241, CNR2-selective agonist, 10 µM) cannabinoids during HSC formation (12–30 hpf) increased runx1 expression in the AGM (n e 90 per condition). (B): In vivo imaging of flk1:dsRed;cmyb:egfp embryos indicated the number of Flk1:dsRed+;cMyb:GFP+ HSCs (arrowheads) was increased in the AGM at 36 hpf following exposure to endogenous and synthetic cannabinoid compounds (n e 7 per condition). (C): Qualitative phenotypic distribution of embryos from panel (A) and Supporting Information Figure S1A scored with low, medium, or high runx1 expression in the AGM (O2545: dual CNR1/2 agonist, 5 µM, ACEA: CNR1-selective agonist, 5 µM). (D): Absolute counts of Flk1:dsRed+cMyb:GFP+ HSCs in embryos from panel (B) and Supporting Information Figure S1B (DMSO: 5.4 ± 0.5, 2-AG: 7.3 ± 1.7, AEA: 7.5 ± 2.4, O2545: 7 ± 1.5, ACEA: 6.2 ± 1.2, AM1241: 7.9 ± 1.9; *, p d .05; **, p d .01; ****, p d .0001, two-tailed t test). (E): Whole-mount in situ hybridization of wild type embryos at 18 and 24 hpf showed cnr2 was expressed throughout the embryo and enriched in the AGM region at the time of HSC specification. (F): MO-mediated knockdown of cnr1 or cnr2 revealed that CNR2, but not CNR1, was required for normal runx1 expression (n ≥ 150 per condition). (G): Qualitative phenotypic distribution of embryos from panel (F) scored with low, medium, or high runx1 expression in the AGM. (H): In vivo imaging confirming the number of Flk1:dsRed+;cMyb:GFP+ HSCs (arrowheads) was decreased in the AGM at 36 hpf following cnr2, but not cnr1, MO-mediated knockdown (n ≥ 10 per condition). (I): Absolute counts of Flk1:dsRed+cMyb:GFP+ HSCs from embryos in panel (B) (uninjected: 5.3 ± 0.7, cnr1 MO: 5.3 ± 0.3, cnr2 MO: 3.6 ± 0.3; *, p < .05, two-tailed t test). Scale bars: (A, B, F, H) = 100 µm, (E) =500 µm. Abbreviations: 2AG, 2-arachidonoylglycerol; AEA, anandamide; AGM, aorta-gonad-mesonephros; MO, morpholino. EXPRESSION / LABELING:
PHENOTYPE:
|
|
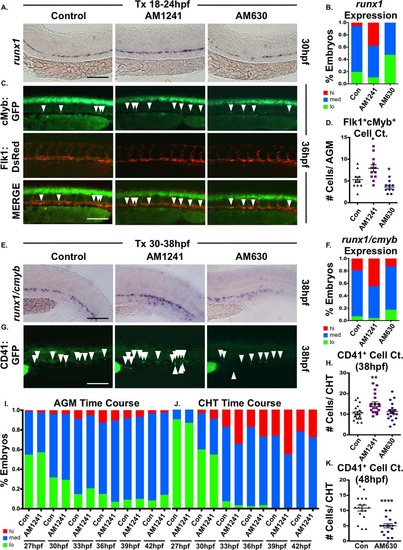
CNR2 modulation impacts hematopoietic stem cell (HSC) development in the AGM and CHT. (A): Embryos exposed to AM1241 or AM630 (CNR2-selective antagonist, 10 µM each) during niche specification (18–24 hpf) exhibited increased or decreased runx1 expression in the AGM at 30 hpf, respectively (n e 55 per condition). (B): Qualitative phenotypic distribution of embryos from panel (A), scored with low, medium, or high runx1 expression in the AGM. (C): In vivo imaging confirmed the number of Flk1:dsRed+;cMyb:GFP+ HSCs (arrowheads) in the AGM was altered at 36 hpf following exposure (18–24 hpf) to AM1241 or AM630 (n e 10 per condition). (D): Absolute counts of Flk1:dsRed+cMyb:GFP+ HSCs from embryos in panel (C) (DMSO: 5.3 ± 0.7, AM1241: 7.9 ± 0.8, AM630: 3.6 ± 0.4; *, p d .05, two-tailed t test). (E): Embryos exposed to CNR-modifiers during the phase of HSC production (30–38 hpf) exhibited altered runx1;cmyb+ expression in the CHT (scored as the vascular region beyond the tip of the yolk sac extension) at 38 hpf (n ≥ 55 per condition). (F): Qualitative phenotypic distribution of embryos from panel (E), scored with low, medium, or high runx1;cmyb expression in the CHT. (G): In vivo imaging of cd41:egfp embryos indicated the number of CD41+ HSCs (arrowheads) was increased in the CHT at 38 hpf following exposure to AM1241 but not significantly altered in embryos exposed to AM630 (n ≥ 20 per condition). (H): Absolute counts of CD41:GFP+ cells from embryos in panel (G) (DMSO: 10.6 ± 0.8, AM1241: 15.0 ± 1.1, AM630: 11.3 ± 0.9; **, p d .01, two-tailed t test). (I): Time course analysis of the qualitative phenotypic distribution of runx1;cmyb expression in the AGM region of embryos exposed to AM1241 during the phase of HSC production (24 hpf until time indicated) (n ≥ 55 per condition). (J): Time course analysis of the qualitative phenotypic distribution of runx1;cmyb expression in the CHT region as described for panel (I), showing increased colonization in response to AM1241 compared to wild type sibling controls (see also Supporting Information Fig. S2E). (K): Embryos exposed to AM630 during HSC production (30–48 hpf) had decreased numbers of CD41+ HSCs at 48 hpf (DMSO: 11.6 ± 0.6, AM630: 7.2 ± 0.6; ****, p ≤ .0001, two-tailed t test; see also Supporting Information Fig. S2F). Scale bars (A, C, E, G) = 100 µm. Abbreviations: AGM, aorta-gonad-mesonephros; CHT, caudal hematopoietic tissue. |
|
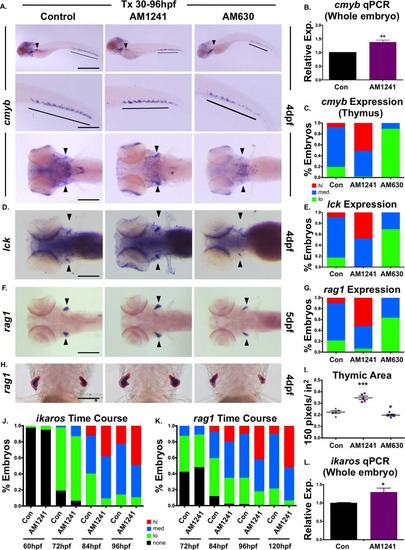
CNR2 modulation impacts hematopoietic stem and progenitor cell colonization of thymus. (A): Exposure to CNR2-modulators (30–96 hpf) during the window of hematopoietic stem cell (HSC) expansion and secondary niche colonization affected cmyb+ expression (upper row, lateral view; line, caudal hematopoietic tissue (CHT) region; arrowhead, kidney marrow; middle row, close up of CHT; lower row, dorsal view; arrowheads, thymi) with AM1241 increasing thymic colonization, and AM630 decreasing expression in all hematopoietic sites (n > 45 per condition). (B): qPCR analysis indicated AM1241-treatment (30–96 hpf) increased total cmyb expression at 4 dpf (n = 4, AM1241: 1.38 ± 0.07-fold; p ≤ .01, two-tailed t test). (C): Qualitative phenotypic distribution of low, medium, or high thymic cmyb expression from embryos exposed to AM1241 and AM630 (30–96 hpf) as in panel (A). (D): Embryos exposed to AM1241 and AM630 during HSC expansion as in panel (A) showed an altered thymic expression of lck (n > 20 per condition). (E): Qualitative phenotypic distribution of embryos from panel (D), scored with low, medium, or high lck expression in the thymus. (F): Embryos exposed to AM1241 and AM630 as in panel (A) showed increased or decreased thymic expression of rag1, respectively (n > 45 per condition). (G): Qualitative phenotypic distribution of embryos from panel (F), scored with low, medium, or high rag1 expression in the thymus. (H): Representative examples of total thymic area measurements (outlined in red) in AM1241- or AM630-exposed embryos (30–96 hpf) following rag1 whole mount in situ hybridization. (I): Bar graph of total thymic area from panel (H) (DMSO: 0.224 ± 0.012 a.u., AM1241: 0.347 ± 0.015, AM630: 0.197 ± 0.008; *, p ≤ .05; ***, p ≤ .001, one-tailed t test, n = 5). (J): Time course analysis (60–96 hpf) of the qualitative phenotypic distribution of ikaros expression in the thymus as described for panel (A), showing increased colonization in response to AM1241 compared to wild type sibling controls (n ≥ 40). (K): Time course analysis (72–120 hpf) of the qualitative phenotypic distribution of rag1 expression in the thymus as described for panel (A), indicating increased lymphoid commitment in response to AM1241 compared to controls (n ≥ 40). (L): qPCR confirmation of increase ikaros expression at 4 dpf in embryos exposed to AM1241 during HSC expansion and niche colonization (30–96 hpf) (AM1241: 1.30 ± 0.11-fold, p ≤ .05; *, p ≤ .05, two-tailed t test, n = 3). Scale bars (A, top) = 800 µm, (A, middle) = 400 µm, (A, bottom), (D, F) = 200 µm, (H) = 100 µm. Abbreviation: qPCR, quantitative polymerase chain reaction. |
|
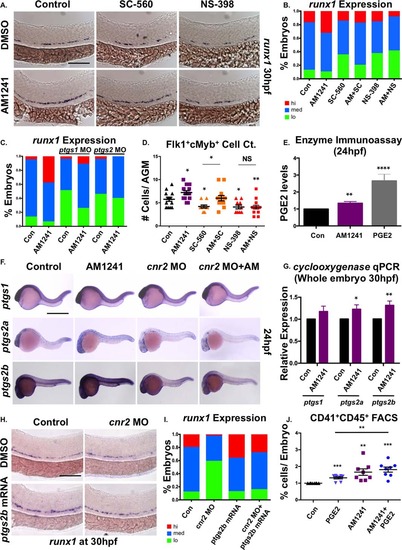
CNR2-signaling increases PGE2 production via ptgs2 induction. (A): Treatment with selective cyclooxygenase (Ptgs)-1 (SC-560, 10 µM) and Ptgs2 (NS-398, 10 µM) inhibitors (12–30 hpf) decreased runx1 expression in the AGM; the effect of SC-560 on runx1, but not that of NS-398, was ameliorated by addition of AM1241 (n ≥ 90 per condition). (B): Qualitative phenotypic distribution of embryos from panel (A), scored with low, medium, or high runx1 expression in the AGM. (C): Qualitative phenotypic distribution of embryos injected with ptgs1 or ptgs2a/ptgs2b MOs in the presence or absence of AM1241-treatment, scored with low, medium, or high runx1 expression; the effect of MO-mediated reduction in Ptgs1, but not that of Ptgs2, on runx1 was restored to wild type levels by exposure to AM1241 (n ≥ 25 per condition). (D): Quantification of the independent and combined effects of selective Ptgs inhibitors and AM1241 by absolute counts of Flk1:dsRed+;cMyb:GFP+ hematopoietic stem cells in embryos, treated as in panel (A), indicated AM1241 acts via Ptgs2 to impact HSCs (DMSO: 5.8 ± 0.5, AM1241: 7.2 ± 0.4, SC-560, 4.1 ± 0.4, AM1241+SC-560: 6.0 ± 0.6, NS-398: 05.8 ± 0.5, AM1241+NS-398: 04.1 ± 0.4; *, p ≤ .05, two-tailed t test, n ≥ 10 per condition). (E): Embryos exposed to AM1241 (12–24 hpf) exhibited augmented PGE2 production: relative concentration of PGE2 was measured in AM1241- and PGE2-treated (positive control) embryos via a biochemical assay for PGE2-metabolites (PGE2, 2.66-fold; AM1241, 1.35-fold; **, p < .01, two-tailed t test, n = 7). (F): Whole-mount in situ hybridization for ptgs1, ptgs2a, and ptgs2b at 24 hpf showed each was upregulated in response to AM1241 exposure (12–24 hpf); MO knockdown of cnr2 decreased the expression level of ptgs2a and ptgs2b, but not of that of ptgs1, and blocked the transcriptional response to AM1241-treatment (n e 25 per condition). (G): qPCR analysis at 30 hpf confirmed ptgs2a and ptgs2b were significantly upregulated over baseline following AM1241 exposure, whereas ptgs1 expression was not (ptgs1: 1.17-fold, NS; ptgs2a: 1.22-fold, *, p < .05; ptgs2b: 1.31-fold, **, p d .01, two-tailed t test, n = 25 pooled embryos per condition × >10 replicates). (H): Overexpression of ptgs2b normalized runx1 expression in the AGM of cnr2 morphants (n e 40 per condition). (I): Qualitative phenotypic distribution of embryos from panel (I), scored with low, medium, or high runx1 expression in the AGM. (J): The effect of combinatorial AM1241- and dmPGE2-treatment (5 µM each) was quantified by FACS using cd41:gfp;cd45:dsRed embryos (dmPGE2: 1.32-fold, AM1241: 1.66-fold, AM1241+dmPGE2: 1.82-fold; **, p d .01; ***, p d .001, two-tailed t test, n = 8 per condition). Scale bars (A) = 100 µm, (F) = 800 µm, (H) = 125 µm. Abbreviations: AGM, aorta-gonad-mesonephros; DMSO, Dimethyl Sulfoxide; FACS, fluorescence-activated cell sorting; MO, morpholino; PGE2, prostaglandin E2; ptgs, prostaglandin endoperoxide synthase; qPCR, quantitative polymerase chain reaction. |
|
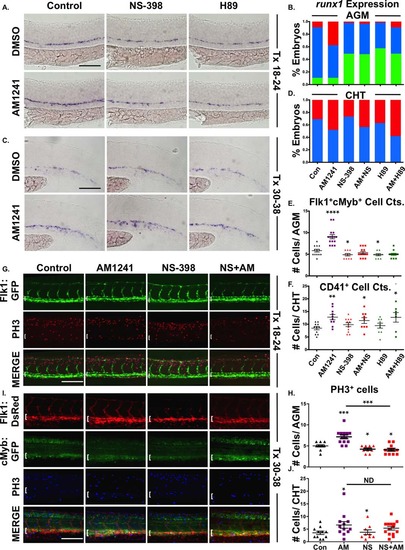
AM1241 mediates hematopoietic stem cell (HSC) expansion in the AGM, but not CHT, via PGE2-signaling. (A): Treatment with selective inhibitors to Ptgs2 (NS-398) or PKA/cAMP (H89, 5 µM) during niche specification (18–24 hpf) decreased runx1 expression and blocked AM1241-mediated inductions in hematopoietic stem and progenitor cell (HSPC) expression in the AGM (n e 45 per condition). (B): Qualitative phenotypic distribution of embryos from panel (A), scored with low, medium, or high runx1 expression in the AGM. (C): Treatment with NS-398 or H89 during hematopoietic stem cell (HSC) production (30–38 hpf) failed to block AM1241-mediated elevations in runx1;cmyb expression in the CHT (n e 75 per condition). (D): Qualitative phenotypic distribution of embryos from panel (C), scored with low, medium, or high runx1 expression in the CHT. (E): Absolute counts of Flk1:dsRed+;cMyb:GFP+ HSCs confirmed PGE2 production and signaling are required to mediate the effects of AM1241 in the AGM (DMSO: 5.8 ± 0.3, AM1241: 9.1 ± 0.5, NS-398: 4.9 ± 0.3, AM1241+NS-398: 5.4 ± 0.3, H89: 4.9 ± 0.2, AM1241+NS-398: 5.0 ± 0.2; *, p d .05; ****, p < .0001, two-tailed t test, n e 10 per condition). (F): Absolute counts of CD41:GFP+ HSCs confirmed AM1241-mediated increases in the CHT are independent of PGE2 production and signaling (DMSO: 8.5 ± 0.6, AM1241: 12.8 ± 1, NS-398: 9.8 ± 0.8, AM1241+NS-398: 11.4 ± 1.1, H89: 9.4 ± 1.0, AM1241+NS-398: 12.7 ± 1.9, *, p d .05; **, p d .01, two-tailed t test, n e 9 per condition). (G): Embryos exposed to AM1241 during niche specification (18–24 hpf) exhibited an increased number of pH3+ (red) cells in the Flk1:GFP+ hemogenic vasculature of the AGM (dorsal aorta marked with white bracket) at 30 hpf, which was blocked by cotreatment with NS-398 (n e 9 per condition). (H): Absolute counts of pH3+ cells in the AGM, as described in panel (G) (DMSO: 5.0 ± 0.27, NS-398: 4.22 ± 0.28, AM1241: 7.15 ± 0.45, AM1241+NS-398: 4.09 ± 0.34; *, p d .05; ***, p d .001, two-tailed t test). (I): Embryos exposed to AM1241 during HSC production (30–38 hpf) exhibited a higher number of pH3+ (blue) cMyb:GFP+ HSPCs in the Flk1:dsRed vasculature of the CHT (CHT width marked with white bracket) at 38 hpf; this effect was independent of Ptgs2 activity (n e 9 per condition). (J): Absolute counts of cMyb:GFP+;pH3+ cells in the CHT, as described in panel (I) (DMSO: 3.55 ± 0.57, NS-398: 3.78 ± 0.94, AM1241: 6.67 ± 1.33, AM1241+NS-398: 5.36 ± 0.72; *, p < .05, two-tailed t test). Scale bars (A, C) = 80 µm, (G, I) = 100 µm. Abbreviations: AGM, aorta-gonad-mesonephros; CHT, caudal hematopoietic tissue; DMSO, Dimethyl Sulfoxide; GFP, green fluorescent protein; PH3, phospho-histone 3. |
|
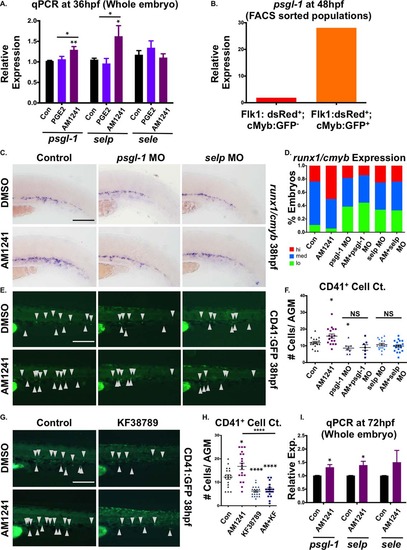
P-selectin mediates the effect of CNR2-stimulation in the caudal hematopoietic tissue (CHT). (A): qPCR analysis revealed expression of p-selectin (selp) and its ligand (psgl-1) were significantly upregulated by AM1241- but not PGE2-exposure (30–36 hpf) (psgl-1: dmPGE2, 1.06-fold; AM1241, 1.29-fold; selp: dmPGE2, 0.96-fold; AM1241, 1.62-fold; sele: dmPGE2, 1.34-fold; AM1241, 1.10-fold; *, p d .05; **, p d .01, two-tailed t test, n e 7). (B): qPCR analysis using FACS-sorted populations from flk1:dsRed;cmyb:GFP embryos at 48 hpf showed that psgl-1 is expressed at a higher level in HSCs (Flk1:dsRed+;cMyb:GFP+: 28.1 a.u.) than in the vasculature (Flk1:dsRed+; cMyb:GFP: 1.86 a.u.) (normalized to 18s). (C): Knockdown of psgl-1 and selp by MO injection reduced runx1;cmyb expression in the CHT and inhibited the effect of AM1241 (30–38 hpf) on hematopoietic stem and progenitor cell production (n e 60 per condition). (D): Qualitative phenotypic distribution of embryos from panel (C), scored with low, medium, or high runx1;cmyb expression in the CHT at 38 hpf. (E): In vivo imaging of cd41:egfp embryos showed that P-selectin knockdown decreased the number of CD41:GFP+ HSCs in the CHT (arrowheads) at 38 hpf and prevented AM1241-stimulated expansion (n e 7 per condition). (F): Absolute counts of CD41:GFP+ cells confirmed P-selectin activity was required for increased hematopoietic stem cell (HSCs) numbers in the CHT mediated by AM1241 (control/DMSO: 11.9 ± 0.7, psgl-1 MO/DMSO: 8.6 ± 1.1, selp MO/DMSO: 9.9 ± 0.7 control/AM1241: 15.8 ± 1.2, psgl-1 MO/AM1241: 9.0 ± 1.7, selp MO/AM1241: 10.7 ± 0.8; *, p d .05, two-tailed t test). (G): Exposure (30–38 hpf) to a selective inhibitor of P-selectin-mediated cell adhesion (KF38789, 0.5 µM) confirmed the requirement of P-selectin activity for normal and AM1241-enhanced CD41:GFP+ HSC numbers in the CHT (n e 18). (H): Absolute counts of CD41:GFP+ HSCs in the CHT after exposure to KF38789 and/or AM1241 (DMSO: 12.2 ± 1.0, AM1241: 16.8 ± 1.4, KF38789: 6.2 ± 0.5, AM1241+KF38789: 6.9 ± 0.7; *, p d .05; ****, p d .0001, two-tailed t test). (I): qPCR analysis showed that expression of selp and psgl-1 was significantly upregulated during HSC expansion and secondary niche colonization (30–72 hpf) (psgl-1: 1.31-fold; selp: 1.40-fold; sele: 1.51-fold; *, p d .05, two-tailed t test, n = 4 replicates). Scale bars (C, E, G) = 100 µm. Abbreviations: DMSO, Dimethyl Sulfoxide; FACS, fluorescence-activated cell sorting; MO, morpholino; PGE2, prostaglandin E2; psgl-1, p-selectin glycoprotein ligand-1; qPCR, quantitative polymerase chain reaction; selp, p-selectin; sele, e-selectin. |
|
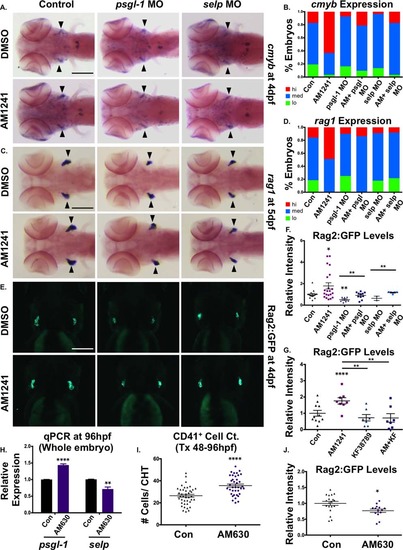
CNR2-signaling promotes thymic colonization via P-selectin activity. (A): MO-mediated knockdown of psgl-1 and selp reduced cmyb expression in the thymus and prevented AM1241 (30–96 hpf) from increasing thymic colonization (n e 50 per condition). (B): Qualitative phenotypic distribution of embryos from panel (A), scored with low, medium, or high cmyb expression in the thymus at 96 hpf. (C): MO-mediated knockdown of psgl-1 and selp, as described in panel (A), decreased rag1 expression in the thymus in the presence and absence of AM1241 (30–120 hpf) (n e 35 per condition). (D): Qualitative phenotypic distribution of embryos from panel (C), scored with low, medium, or high rag1 expression in the thymus at 96 hpf. (E): In vivo imaging of rag2:egfp embryos confirmed that psgl-1- and selp-MOs decreased the number of Rag2:GFP+ lymphoid progenitors in the thymus both with and without AM1241-treatment (n e 8 per condition). (F): Relative fluorescent intensity of the thymus in morphant embryos, as described in panel (E) (control/DMSO: 1.00 ± 0.10 a.u., psgl-1 MO/DMSO:0.51 ± 0.08, selp MO/DMSO: 0.64 ± 0.29, Control/AM1241: 1.79 ± 0.29 a.u., psgl-1 MO/AM1241: 0.91 ± 0.09, selp MO/AM1241: 1.17 ± 0.53; *, p d .05; **, p d .01, one-tailed t test). (G): Exposure to KF38789 (30–96 hpf) confirmed the role of P-selectin in normal and AM1241-enhanced thymic colonization, as indicated by Rag2:GFP at 96 hpf (DMSO: 1.00 ± 0.18 a.u., AM1241: 1.76 ± 0.20, KF38789: 0.73 ± 0.20, AM1241+KF38789: 0.71 ± 0.26; **, p d .01; ****, p d .0001, two-tailed t test, n e 7 per condition). (H): qPCR analysis showed that exposure to AM630 after the phase of HSC induction (48–96 hpf) altered the expression of psgl-1 and selp (psgl-1: 1.44-fold, selp: 0.72-fold, **, p d .01; ****, p d .0001, two-tailed t test). (I): Absolute counts of CD41:GFP+ cells revealed that exposure to AM630, as described in panel (H), increased the number of HSCs remaining in the CHT at 96 hpf (DMSO: 26.4 ± 1.3, AM630: 35.6 ± 1.4; ****, p d .0001, two-tailed t test, n e 38,). (J): Relative fluorescent intensity of Rag2:GFP+ cells the thymus following exposure to AM630, as described in panel (G), indicated CNR2-signaling was required for thymic colonization (DMSO: 1.0 ± 0.06 a.u., AM630: 0.77 ± 0.05 a.u.; *, p d .05, two-tailed t test, n e 14). Scale bars (A, C) = 200 µm, (E) = 175 µm. Abbreviations: DMSO, dimethyl sulfoxide; GFP, green fluorescent protein; MO, morpholino; psgl-1, p-selectin glycoprotein ligand-1; qPCR, quantitative polymerase chain reaction; sele, e-selectin. |
|
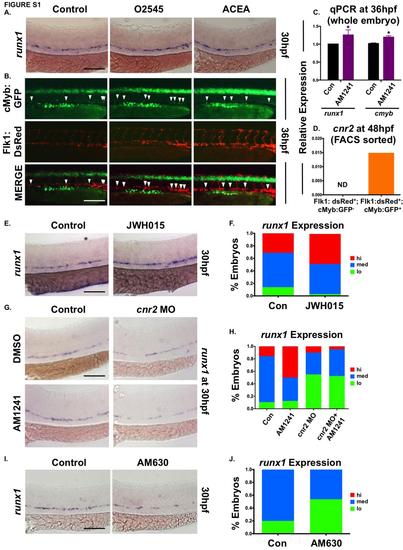
CNR2-selective signaling modulates HSC number in the AGM. (A) Exposure of WT embryos to a synthetic dual CNR1-/CNR2-selective agonist (O2545) or CNR1- selective agonist (ACEA) during HSC formation (12-30hpf) indicated that only CNR2-stimulation impacted runx1 expression in the AGM (n>85/condition; see also Figure 1B). (B) In vivo imaging of flk1:dsRed;cmyb:egfp embryos showed the number of Flk1:dsRed+;cMyb:GFP+ HSCs (arrowheads) was increased in the AGM at 36hpf following exposure to O2545 but not ACEA (CNR1-selective) (ne7/condition; see also Figure 1D). (C) qPCR quantification of the increase in runx1 and cmyb expression at 36hpf in embryos exposed to AM1241 during HSC formation (12-36hpf) (runx1: 1.25-fold, cmyb: 1.20-fold, *p≤0.05, 1-tailed ttest, n=25 pooled embryos/condition x 4 replicates). (D) qPCR analysis of FACS-sorted populations from flk1:dsRed;cmyb:egfp embryos at 48hpf revealed cnr2 was expressed on HSCs (Flk1:dsRed+ ;cMyb:GFP+) but not on endothelium (Flk1:dsRed+;cMyb:GFP-) (HSC: 0.015 a.u.; endothelium: not detected (ND), normalized to 18s). (E) Exposure to JWH015 (CNR2-selective agonist) during HSC formation (12-30hpf) increased runx1 expression in the AGM (n≥130/condition) at 30hpf. (F) Qualitative phenotypic distribution of embryos from panel S1E, scored with low, medium or high runx1 expression in the AGM. (G) AM1241-exposure failed to increase runx1 expression in the AGM of cnr2 morphants, confirming phenotypic receptor specificity (n≥15/condition) at 30hpf. (H) Qualitative phenotypic distribution of embryos from panel S1G, scored with low, medium or high runx1 expression in the AGM. (I) Exposure to the CNR2 antagonist AM630 during HSC formation (12-30hpf) reduced runx1 expression in the AGM (n≥50/condition) at 30hpf. (J) Qualitative phenotypic distribution of embryos from panel S1I, scored with low, medium or high runx1 expression in the AGM. Scale bars: A,B,E,G,I=100µm. |
|
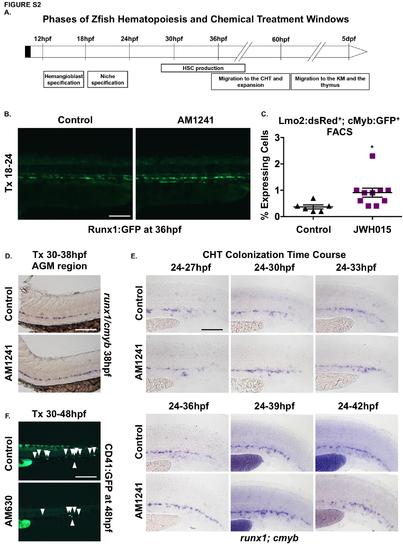
CNR2-signaling is required for HSC production, expansion and CHT colonization. (A) Schematic representation of the different stages of HSC development starting during somitogenesis (12hpf) and continuing until larval stages (5dpf). HSCs are specified in the hemogenic endothelium prior to 24hpf, and are actively produced between 28 and 48hpf in the AGM. HSCs bud and migrate to CHT starting around 33hpf. HSC expansion occurs in the CHT, and HSCs begin to exit to seed the kidney marrow (KM) and thymus by 72hpf. (B) In vivo imaging confirmed numbers of Runx1:GFP+ HSPCs were increased in the AGM at 36hpf following AM1241 exposure during niche specification (18-24hpf) (n≥10/condition). (C) The effect of JWH015 (18-24hpf) on HSPCs was quantified by FACS analysis of cmyb:gfp;lmo2:dsRed2 embryos (2.48-fold; *p<0.05, 2-tailed t-test, n≥6/condition). (D) Embryos exposed to AM1241 during HSC production (30-38hpf) exhibited no apparent increase in runx1;cmyb+ HSPCs within the AGM region (n≥65/condition). (E) Time course analysis (24-42hpf) of the qualitative phenotypic distribution of runx1;cmyb expression in the CHT, indicating increased HSPC colonization in response to AM1241-treatment as compared to controls (ne55/condition). (F) In vivo imaging of cd41:egfp embryos indicated the number of CD41:GFP+ HSCs (arrowheads) was decreased in the CHT at 48hpf following exposure to AM630 (n≥15/condition; see also Figure 2K). Scale bars: B,D,E,F=100µm. |
|
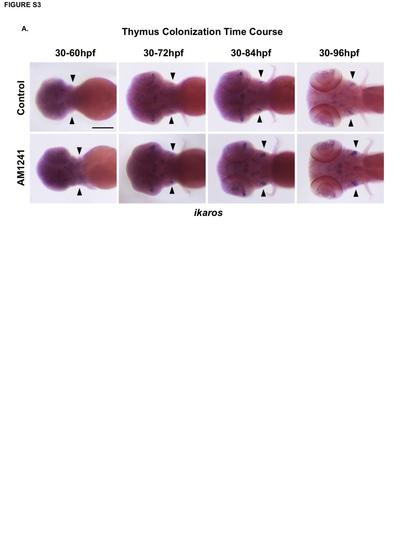
CNR2-signaling increases the number of ikaros+ progenitors in the thymus. (A) Time course analysis of ikaros expression during thymus colonization (60-96hpf) revealed increased thymic progenitors present by 72hpf in embryos exposed to AM1241 compared to controls (n≥40/condition; see also Figure 3J). Scale bars: A=200µm. |
|
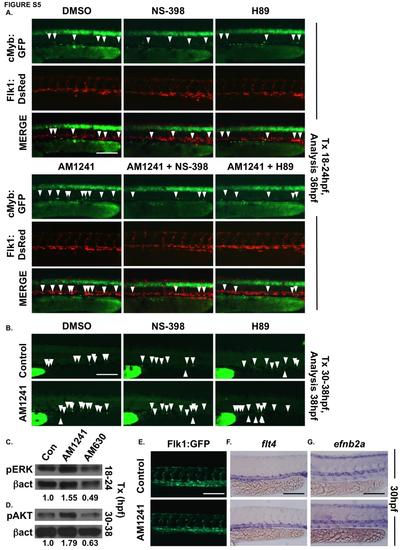
AM1241 increases HSC number in the AGM in a PGE2-dependent manner, without affecting the vasculature. (A) In vivo imaging of flk1:dsRed;cmyb:egfp embryos confirmed PGE2 production and signaling are required to mediate the effect of AM1241 on AGM HSCs (n≥10/condition). (B) In vivo imaging of cd41:gfp embryos confirmed that the AM1241-mediated increase in HSCs in the CHT is independent of PGE2 production and signaling (n≥9/condition). (C) Western blot analysis showed CNR2-modulation during niche specification altered pERK levels at 24hpf (DMSO: 1 a -actin, n=50 pooled embryos/condition, x 3 replicates). (D) Western blot analysis confirmed CNR2-modulation altered pAKT protein levels at 24hpf (DMSO: 1-actin, n=50 pooled embryos/condition x 3 replicates). (E) Flk1:GFP vascular patterning was not impacted by AM1241 exposure (12-30hpf) (n≥20/condition). (F) Expression of flt4 in the vein was not affected in AM1241-treated embryos (n≥25/condition). (G) Arterial identity, as indicated by ephrinB2a, was not altered in AM1241-exposed embryos (n≥20/condition). Scale bars: A,B,G=100µm, E,F=125µm. |
|
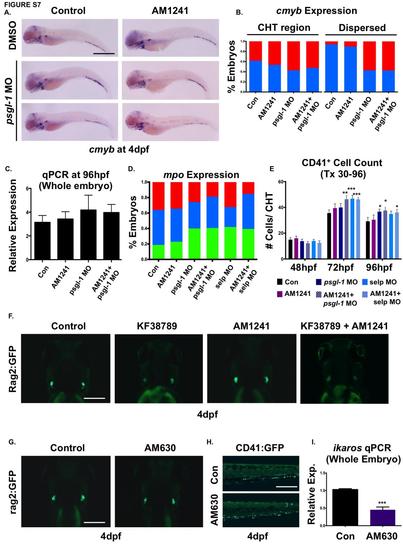
CNR2-signaling promotes HSPC migration to secondary niches via P-selectin. (A) In psgl-1 morphant embryos, while thymic colonization is reduced, cmyb expression is observed to accumulate in the CHT (middle panel) and/or appear in isolated cells scatter throughout the embryo (lower panel) in the presence and absence of AM1241-treatment (30-96hpf). (B) Qualitative phenotypic distribution of cmyb expression within the CHT (left side) or dispersed throughout the embryo (right side) in psgl-1 morphants with and without AM1241-treatment compared to sibling controls (n≥35/condition). (C) qPCR analysis of psgl-1 morphant embryos showed no significant alterations in cmyb expression in the presence or absence of AM1241- : 1.09-fold, psgl-1 MO/DMSO : 1.33-fold, psgl-1 MO/AM1241 : 1.26-fold; NS, 2-tailed t-test, n=25 pooled embryos/condition x 4 replicates). (D) Qualitative phenotypic distribution of embryos injected with psgl-1 and selp MOs and subsequently exposed to AM1241 revealed mpo expression, indicative of myeloid commitment, was not increased following P-selectin knockdown (n≥30/condition). (E) Absolute counts of CD41:GFP+ cells from embryos injected with psgl-1 and selp MOs and exposed to AM1241 (30-96hpf, 5µM) revealed inhibition of P-selectin activity caused prolonged retention of HSCs in the CHT compared to controls (48hpf: Uninjected/DMSO: 15±1.5, Uninjected/AM1241:16.1±1.7, psgl-1 MO/DMSO: 13.8±1.5, psgl-1 MO/AM1241:12.2±1.3, selp MO/DMSO: 14.0±1.2, selp MO/AM1241: 12.6±1.5; 72hpf: Uninjected/DMSO: 35.6±2.4, Uninjected/AM1241:39.4±3.4, psgl-1 MO/DMSO: 39.9±3.1, psgl-1 MO/AM1241:46.4±3.2, selp MO/DMSO: 46.7±2.7, selp MO/AM1241: 46.2±2.0; 96hpf: Uninjected/DMSO: 29.2±3.1, Uninjected/AM1241:30.5±3.7, psgl-1 MO/DMSO: 36.7±1.9, psgl-1 MO/AM1241:37.6±3.3, selp MO/DMSO: 34.9±2.3, selp MO/AM1241: 36.1±2.6; *p≤0.05, **p≤0.01, ***p≤0.001, 2-tailed t-test, n≥10/condition). (F) Embryos exposed to KF38789 (30-96hpf) confirmed the effects of P-selectin on normal and AM1241- enhanced colonization of Rag2:GFP+ lymphoid progenitors in the thymus at 4dpf (n≥10/condition). (G) Embryos exposed to AM630 during HSC expansion and thymic colonization (48-96hpf) showed decreased Rag2:GFP intensity in the thymus (n≥10/condition). (H) In vivo imaging of cd41:gfp embryos (48-96hpf) showed exposure to AM630 cause HSC numbers to remain elevated in the CHT at 96hpf (n≥15/condition). (I) qPCR analysis confirmed decreased ikaros expression at 96hpf in embryos exposed to AM630 (30- 96hpf) (0.65-fold; ***p≤0.001, 2-tailed t-test, n=25 pooled embryos/condition x 4/replicates). Scale bars: A =800µm, F,G=175µm, H=200µm. |