- Title
-
Live imaging of endogenous Collapsin response mediator protein-1 expression at subcellular resolution during zebrafish nervous system development
- Authors
- Jayasena, C.S., and Bronner-Fraser, M.
- Source
- Full text @ Gene Expr. Patterns
|
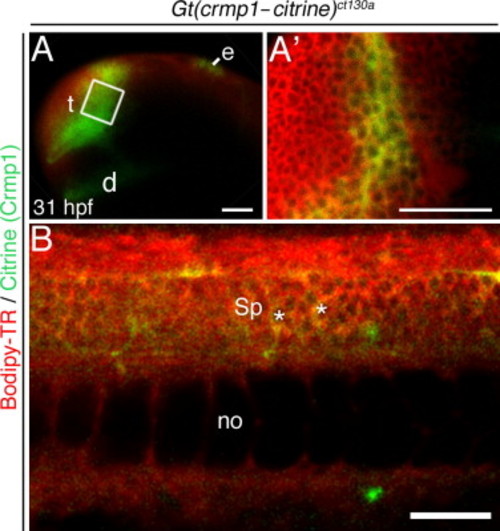
Crmp1 (green) is expressed in the forebrain at 31 hpf. All embryos were treated with the vital dye Bodipy-TR (red) to act as a contrast against the citrine signal. (A–B) Live confocal images. (A) Confocal z-stack showing Citrine/ Crmp1 (green) expression in the forebrain regions: the telencephalon (t) and diencephalon (d). Also, expression in epiphysis (e) is apparent. Boxed area shown in (A2). (A2) Z-section through the telencephalon. (B) Expression also found in spinal cord neurons (Sp) (asterisks). no, notochord. Scale bar: 50 μm. EXPRESSION / LABELING:
|
|
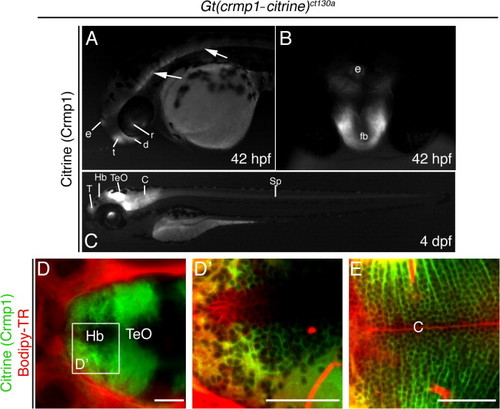
Crmp1 expression in the developing brain in live 42 hpf- 4 day fish. (A) Lateral view at 42 hpf. Crmp1 is expressed in the epiphysis (e) and forebrain regions: telencephalon (t) and diencephalons (d). Expression is also detected in the retina (r). Arrows show expression in brain and spinal cord. (B) Frontal view at 42 hpf of the forebrain (fb) and epiphysis. (C) Expression in the 4 day live larva showing expression in the T, telecephalon, Hb, habenula, TeO, optic tectum and Cb, cerebellum. Expression in spinal cord (Sp) also persists. (D–E) Larvae were treated with the vital dye Bodipy-TR (red) to act as a contrast against the citrine signal. (D) Confocal z-stack at 4 dpf, dorsal view showing expression in in Hb and TeO. (D2) Shows zoomed view but z-section. (E) Z-section at the level of cerebellum. Scale bar: 50 μm. |
|
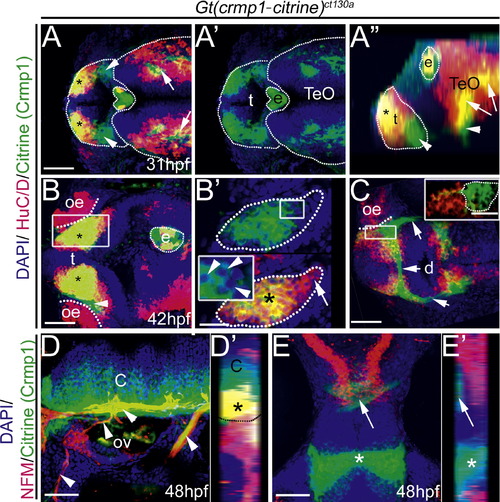
Crmp1 expression (green) compared to neuronal markers HuC/D (A–C) and NFM (D–E) (red). Gt(crmp1-citrine)ct130a heterozygotes were stained with anti-green fluorescent protein to enhance the citrine signal and the nuclear stain (DAPI; blue) to visualize nuclei. (A)–(B) Dorsal view z-stack projection at the level of telencephalon (t) and optic tectum (TeO) at 31 hpf (A) and 42 hpf (B). Asterisk: co-expression of HuC/D and Crmp1. e; epiphysis. (A) Arrowheads: Neurons positive for Crmp1 in the telencephalon. Arrows: Crmp1 and HuC/D positive cells in the optic tectum (TeO). (A2) Crmp1 expression alone. (A3) Lateral reconstructed projection of the z-stack from (A). Arrowhead: Crmp1 expression devoid of HuC/D expression. Arrows: Crmp1 and HuC/D positive cells. Asterisk: HuC/D and Crmp1 positive cells in the telencephalon. (B)–(C) oe, olfactory epithelium. (B2) Magnified z-section from boxed region in (B) showing that majority of HuC/D cells are also positive for Crmp1 (highlighted dotted region; asterisk). Arrow: HuC/D positive and Crmp1 negative cells. Inset in (B2) shows that Crmp1 is localized to the cytoplasm (arrowheads) and excluded from the nucleus. (C) Ventral view z-stack projection of the diencephalon (d) at 42 hpf. Arrows: cell projections positive for Crmp1. Inset: magnified view of boxed region. Highlighted Crmp1 cells are negative for HuC/D. (D) Lateral view z-projection at the level of the otic vesicle (ov) and cerebellum (c). Arrowheads: axon projections that are NFM and Crmp1 positive. (E) Crmp1 cells at the level for the forebrain (asterisk) and optic tectum (arrow) are negative for NFM. (D2), (E2) Ninty-degree rotated z-stack reconstructed projection of (D) and (E) respectively. (D2) Dotted outline delineates cerebellum (C) from the peripheral tissue. Asterisk: NFM and Crmp1 positive axon tracks. Scale: 50 μM (A)–(B), (C)–(D), 25 μM ((B2) and inset (C)). |
|
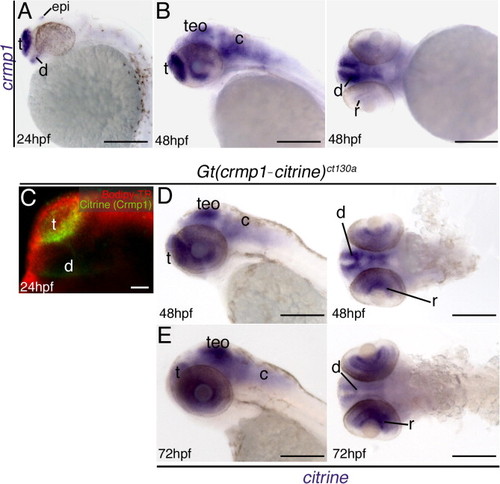
In situ hybridization results showing expression of crmp1 (A)–(B) and citrine (D)–(E). In wild type fish crmp1 expression is localized to the nervous system at 24 hpf (A) and 48 hpf (B). (B) Lateral and frontal views at 48 hpf. (C) Confocal z-stack at 24 h post fertilization (hpf) showing Citrine/ Crmp1 (green) expression in the forebrain regions. Red: Bodipy-TR vital dye. (D)–(E) Lateral and ventral views at 48 hpf and 72 hpf. Transcripts for citrine are expressed in selected neural regions at both stages consistent with wild type expression. Abbreviations: epi, epiphysis; t, telencephalon; teO, optic tectum/midbrain; c, cerebellum/ hindbrain; d, diencephalon; r, retina. Scale: 200 μM, 50 μM (C). EXPRESSION / LABELING:
|
Reprinted from Gene expression patterns : GEP, 11(7), Jayasena, C.S., and Bronner-Fraser, M., Live imaging of endogenous Collapsin response mediator protein-1 expression at subcellular resolution during zebrafish nervous system development, 395-400, Copyright (2011) with permission from Elsevier. Full text @ Gene Expr. Patterns