- Title
-
MicroRNAs and micromanaging the skeleton in disease, development and evolution
- Authors
- He, X., Eberhart, J.K., and Postlethwait, J.H.
- Source
- Full text @ J. Cell. Mol. Med.
|
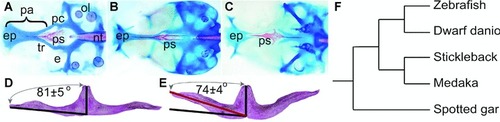
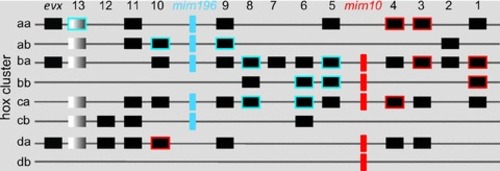
Closely related species and populations within a species often display subtle morphological differences in skeletal systems. Such morphological variation probably results from slight differences in the timing or location or intensity of the action of specific genes. ( |
|
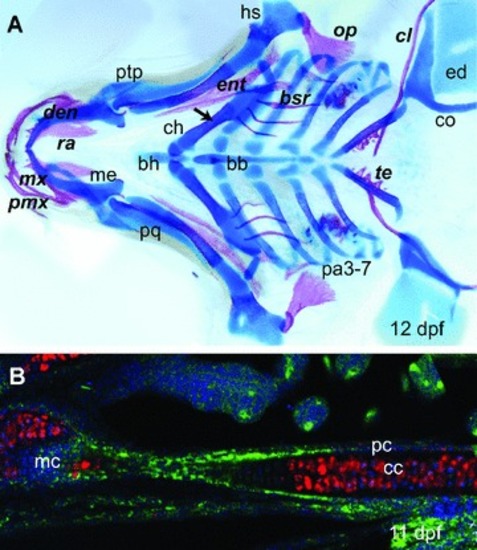
Endochondral ossification and intramembranous ossification. ( |
|
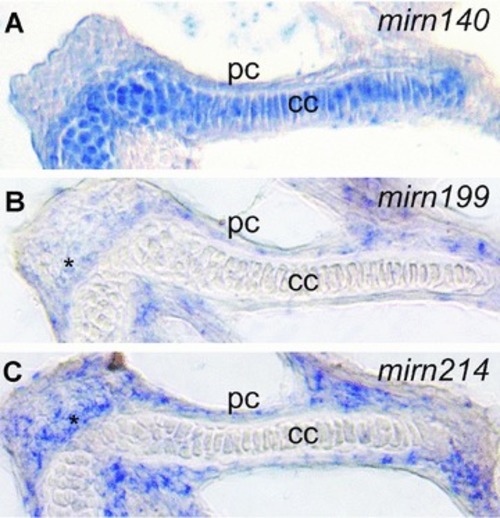
Skeletal miRNAs are expressed in discrete patterns. Conventional |
|
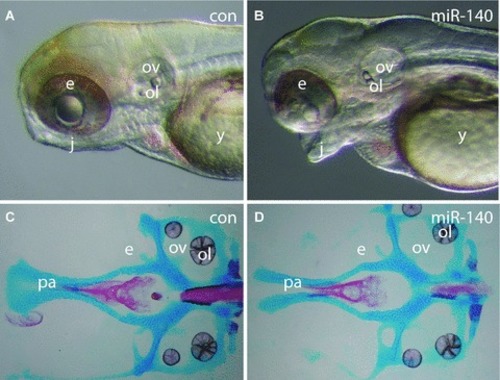
Overexpression of miR-140 causes cleft palate. ( |
|
|
|
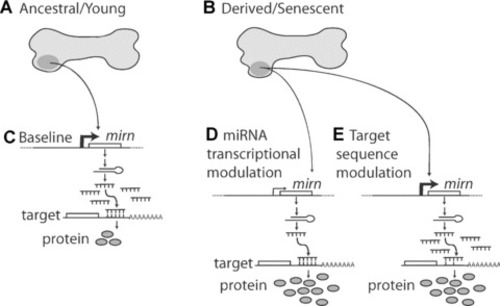
A model for the roles of miRNAs in evolution and disease. ( |