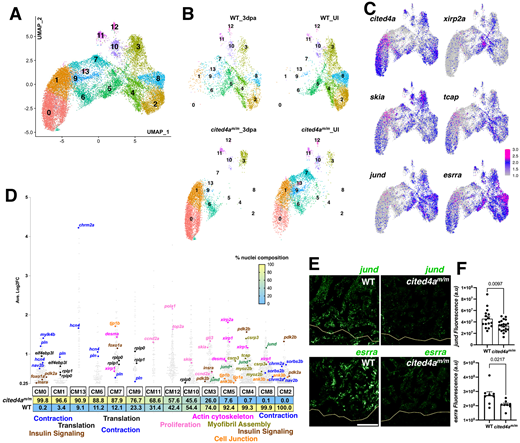
Fig. 8 Second level reclustering and integration of CMs in cited4m/m and WT hearts show distinct CM populations that are not shared. (A,B) UMAP visualization of reclustered CM (A) and split into each condition and genotype (B). (C) Feature plot of gene expression in the CM clusters. (D) Gene expression plot by average log2 fold change in each CM cluster. Each dot represents a single gene, with percentage of nuclei composition for each genotype listed below. (E,F) Representative images of jund and esrra transcripts detected by RNAscope accompanied with graphs showing fluorescent intensity measurements in WT and cited4am/m ventricles at 3 dpa. Data are mean±s.e.m. Unpaired two-tailed t-test. Yellow line in E indicates amputation plane. Fluorescent intensity of RNAscope in F in arbitrary units (a.u.). For jund WT n=4; cited4am/m n=5. For esrra n=4 for each condition. Each dot represents measurements from a single 14 µm section. Four sections per heart for jund, two sections per heart for esrra. Scale bar: 100 µm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development